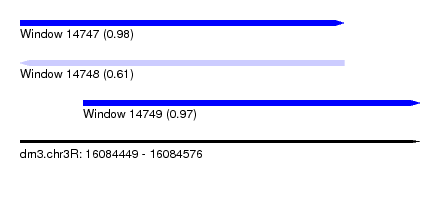
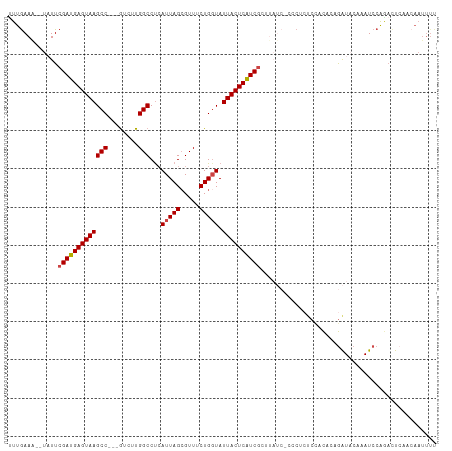
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,084,449 – 16,084,576 |
| Length | 127 |
| Max. P | 0.984366 |

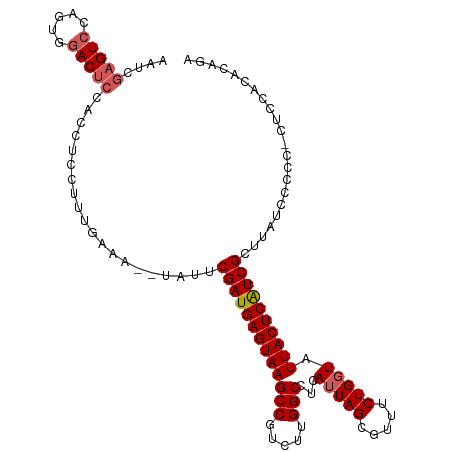
| Location | 16,084,449 – 16,084,552 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.65 |
| Shannon entropy | 0.30824 |
| G+C content | 0.46342 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16084449 103 + 27905053 ------GUCCUGUGGACUCAACCUCCUUUGAAA--UAUUCGAUGAGUAAGCCGUCUUGGCCUAAUUAGCGUUUCUGGUAUUACUCAUCGCUUAUCUCCCACUCCACACAGA ------....((((((.((((......)))).(--((..((((((((((((((....(((((....)).)))..)))).))))))))))..))).......)))))).... ( -26.50, z-score = -2.95, R) >droSec1.super_27 150997 109 - 799419 AAUCGAGUCCAGUGGACUCCACCUCCUUUGAAG--UAUUCGAUGAGUAAGCCGUCUUGGCCUCAUUAGCGUUUCUGGUAUUACUCGUCGCUUAUCCCCCUCUCCACACAGA ....((((((...)))))).............(--((..(((((((((((((.....)))...(((((.....))))).))))))))))..)))................. ( -25.70, z-score = -1.17, R) >droSim1.chr3R 22125038 108 - 27517382 AAUCGAGUCCAGUGGACUCCACCUCCUUUGAAG--UAUUCGAUGAGUAAGCCGUCUUGGCCUCAUUAGCGUUUCUGGUAUUACUCAUCGCUUAUCCCCC-CUCCACACAGA ....((((((...)))))).............(--((..(((((((((((((.....)))...(((((.....))))).))))))))))..))).....-........... ( -26.10, z-score = -1.74, R) >droYak2.chr3R 4864465 95 + 28832112 AAUCGAGUCCAAUGGACUCCACUUCCCUUAAAA--UAUUCGAUGAGUAAGCCGUCUUGGCCUCAUUAGCGUUUCUGGUAUUACUCAUCGCCCACAGA-------------- ....((((((...))))))..............--....(((((((((((((.....)))...(((((.....))))).))))))))))........-------------- ( -24.80, z-score = -2.69, R) >droEre2.scaffold_4770 12154335 109 - 17746568 AAUCGAGUCCAGUAAACUCCACCUCCUUUGAAAAUUUUUCGAUGAGUAAGCCAUCUUGGCCUCAUUAGCGUUUCUGCUAUUACUCAUCGCUUAUCGACCUCUCUACAGA-- ..((((....(((................(((.....)))((((((((((((.....))).....(((((....)))))))))))))))))..))))............-- ( -24.00, z-score = -2.68, R) >consensus AAUCGAGUCCAGUGGACUCCACCUCCUUUGAAA__UAUUCGAUGAGUAAGCCGUCUUGGCCUCAUUAGCGUUUCUGGUAUUACUCAUCGCUUAUCCCCC_CUCCACACAGA ....(((((.....)))))....................(((((((((((((.....)))...(((((.....))))).))))))))))...................... (-21.26 = -21.90 + 0.64)

| Location | 16,084,449 – 16,084,552 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.65 |
| Shannon entropy | 0.30824 |
| G+C content | 0.46342 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -22.04 |
| Energy contribution | -21.76 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16084449 103 - 27905053 UCUGUGUGGAGUGGGAGAUAAGCGAUGAGUAAUACCAGAAACGCUAAUUAGGCCAAGACGGCUUACUCAUCGAAUA--UUUCAAAGGAGGUUGAGUCCACAGGAC------ (((.((((((.(.(((((((..((((((((((...........((....))(((.....)))))))))))))..))--)))).........).).))))))))).------ ( -29.70, z-score = -2.33, R) >droSec1.super_27 150997 109 + 799419 UCUGUGUGGAGAGGGGGAUAAGCGACGAGUAAUACCAGAAACGCUAAUGAGGCCAAGACGGCUUACUCAUCGAAUA--CUUCAAAGGAGGUGGAGUCCACUGGACUCGAUU (((...(((((.........((((.................)))).((((((((.....)))...)))))......--)))))..)))(((.((((((...)))))).))) ( -26.13, z-score = 0.52, R) >droSim1.chr3R 22125038 108 + 27517382 UCUGUGUGGAG-GGGGGAUAAGCGAUGAGUAAUACCAGAAACGCUAAUGAGGCCAAGACGGCUUACUCAUCGAAUA--CUUCAAAGGAGGUGGAGUCCACUGGACUCGAUU (((...(((((-..........((((((((((...........((....))(((.....)))))))))))))....--)))))..)))(((.((((((...)))))).))) ( -30.94, z-score = -1.02, R) >droYak2.chr3R 4864465 95 - 28832112 --------------UCUGUGGGCGAUGAGUAAUACCAGAAACGCUAAUGAGGCCAAGACGGCUUACUCAUCGAAUA--UUUUAAGGGAAGUGGAGUCCAUUGGACUCGAUU --------------...(((..((((((((((...........((....))(((.....)))))))))))))..))--).........(((.((((((...)))))).))) ( -27.10, z-score = -1.70, R) >droEre2.scaffold_4770 12154335 109 + 17746568 --UCUGUAGAGAGGUCGAUAAGCGAUGAGUAAUAGCAGAAACGCUAAUGAGGCCAAGAUGGCUUACUCAUCGAAAAAUUUUCAAAGGAGGUGGAGUUUACUGGACUCGAUU --(((...(((((.........(((((((((.((((......))))....((((.....))))))))))))).....)))))...)))(((.(((((.....))))).))) ( -30.74, z-score = -2.26, R) >consensus UCUGUGUGGAG_GGGCGAUAAGCGAUGAGUAAUACCAGAAACGCUAAUGAGGCCAAGACGGCUUACUCAUCGAAUA__UUUCAAAGGAGGUGGAGUCCACUGGACUCGAUU ......................((((((((((...........((....))(((.....)))))))))))))......((((....))))..((((((...)))))).... (-22.04 = -21.76 + -0.28)

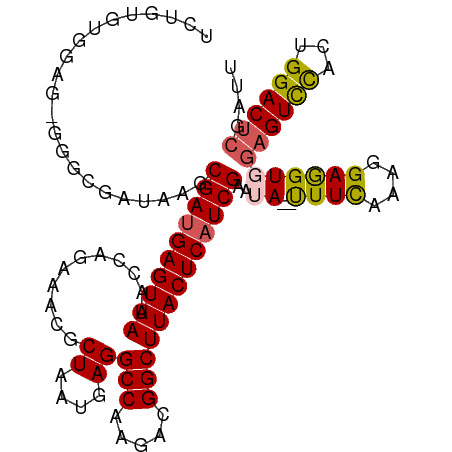
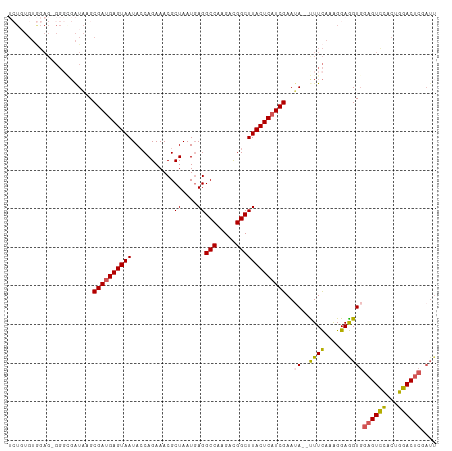
| Location | 16,084,469 – 16,084,576 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.41808 |
| G+C content | 0.42212 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
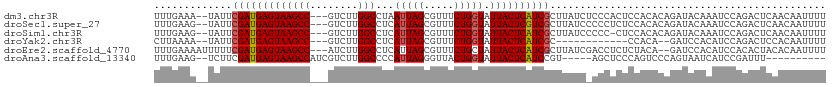
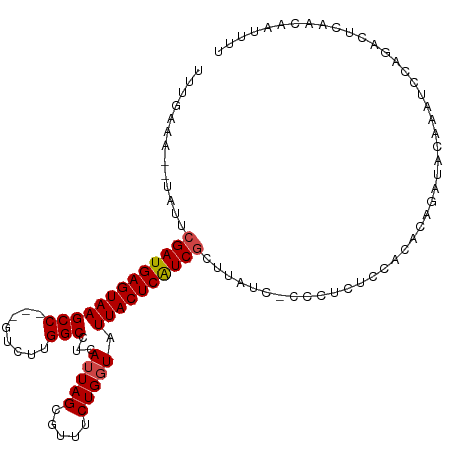
>dm3.chr3R 16084469 107 + 27905053 UUUGAAA--UAUUCGAUGAGUAAGCC---GUCUUGGCCUAAUUAGCGUUUCUGGUAUUACUCAUCGCUUAUCUCCCACUCCACACAGAUACAAAUCCAGACUCAACAAUUUU ...((.(--((..(((((((((((((---(....(((((....)).)))..)))).))))))))))..))).))...................................... ( -19.80, z-score = -1.74, R) >droSec1.super_27 151023 107 - 799419 UUUGAAG--UAUUCGAUGAGUAAGCC---GUCUUGGCCUCAUUAGCGUUUCUGGUAUUACUCGUCGCUUAUCCCCCUCUCCACACAGAUACAAAUCCAGACUCAACAAUUUU .((((.(--((..(((((((((((((---.....)))...(((((.....))))).))))))))))..))).....(((......))).............))))....... ( -18.70, z-score = -0.64, R) >droSim1.chr3R 22125064 106 - 27517382 UUUGAAG--UAUUCGAUGAGUAAGCC---GUCUUGGCCUCAUUAGCGUUUCUGGUAUUACUCAUCGCUUAUCCCCC-CUCCACACAGAUACAAAUCCAGACUCAACAAUUUU .((((.(--((..(((((((((((((---.....)))...(((((.....))))).))))))))))..))).....-.........(((....))).....))))....... ( -19.10, z-score = -1.25, R) >droYak2.chr3R 4864491 93 + 28832112 CUUAAAA--UAUUCGAUGAGUAAGCC---GUCUUGGCCUCAUUAGCGUUUCUGGUAUUACUCAUCGC------------CCACA--GAUCCACAUCCAGACUCCACAAUUUU .......--....(((((((((((((---.....)))...(((((.....))))).)))))))))).------------.....--.......................... ( -17.00, z-score = -1.52, R) >droEre2.scaffold_4770 12154361 107 - 17746568 UUUGAAAAUUUUUCGAUGAGUAAGCC---AUCUUGGCCUCAUUAGCGUUUCUGCUAUUACUCAUCGCUUAUCGACCUCUCUACA--GAUCCACAUCCACACUACACAAUUUU .............(((((((((((((---.....))).....(((((....)))))))))))))))..................--.......................... ( -20.50, z-score = -3.20, R) >droAna3.scaffold_13340 2630283 95 + 23697760 UUUGAAG--UCUUCGAUGAGUAAGCCAUCGUCUUGGCCCCAUUAGGGUUACUGGUAUUACUCAUCCGU-----AGCUCCCAGUCCCAGUAAUCAUCCGAUUU---------- ....(((--((...(((((....((((......)))).......(((..(((((..((((......))-----))...)))))))).....))))).)))))---------- ( -24.00, z-score = -2.08, R) >consensus UUUGAAA__UAUUCGAUGAGUAAGCC___GUCUUGGCCUCAUUAGCGUUUCUGGUAUUACUCAUCGCUUAUC_CCCUCUCCACACAGAUACAAAUCCAGACUCAACAAUUUU .............(((((((((((((........)))...(((((.....))))).)))))))))).............................................. (-15.37 = -15.57 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:25 2011