
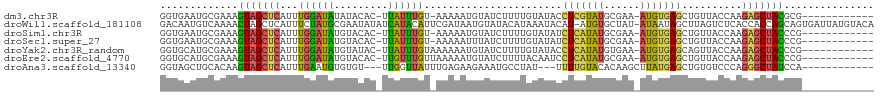
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,035,733 – 16,035,837 |
| Length | 104 |
| Max. P | 0.858091 |

| Location | 16,035,733 – 16,035,837 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 69.12 |
| Shannon entropy | 0.57240 |
| G+C content | 0.36866 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -11.64 |
| Energy contribution | -10.86 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
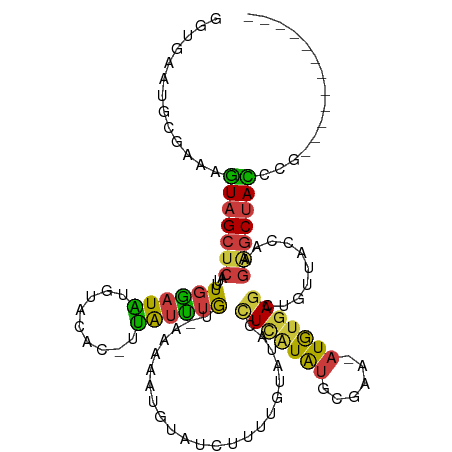
>dm3.chr3R 16035733 104 - 27905053 GGUGAAUGCGAAAGUAGCUCAUUUGGAUAUAUACAC-UUAUUUGU-AAAAAUGUAUCUUUUGUAUACCUCGUAUGCGAA-AUGUGAGCUGUUACCAAGAGCUACGCG------------ (((((.(((....)))((((((..(((((((((((.-.....)))-)....)))))))(((((((((...)))))))))-..))))))..)))))............------------ ( -28.50, z-score = -2.64, R) >droWil1.scaffold_181108 915116 117 + 4707319 GACAAUGUCAAAACUAGCUCAUUUCUAUGCGAAUAUAUCAUACAUUCGAUAAUGUAUACAUAAAUACAU-AUGUGCUAU-AUAAUAGCUUAGUCUCACCACCAGCAGUGAUUAUGUACA .(((.((((.......((..........))((((.........)))))))).))).(((((((......-....(((((-...)))))......((((........))))))))))).. ( -16.90, z-score = 0.70, R) >droSim1.chr3R 22070521 104 + 27517382 GGUGAAUGCGAAAGUAGCUCAUUUGGAUAUGUACAC-UUAUUUGU-AAAAAUGUAUCUUUUGUAUAUCUCAUAUGCGAA-AUGUGAGCUGUUACCAAGAGCUACCCG------------ (((((.(((....)))((((((..((((((.((((.-.....)))-).....))))))(((((((((...)))))))))-..))))))..)))))............------------ ( -26.20, z-score = -2.31, R) >droSec1.super_27 102783 104 + 799419 GGUGAAUGCGAAAGUAGCUCAUUUGGAUAUGUACAC-UUAUUUGU-AAAAAUUUAUCUUUUGUAUAUCUCAUAUGCGAA-AUGUGAGCUGUUACCAAGAGCUACCCG------------ (((((.(((....)))((((((..(((((..((((.-.....)))-)......)))))(((((((((...)))))))))-..))))))..)))))............------------ ( -25.20, z-score = -2.26, R) >droYak2.chr3R_random 2649233 105 - 3277457 GGUGCAUGCGAAAGUAGCUCAUUUGGAUAUGUAUAC-UUAUUUGUAAAAAAUGUAUCUUUUGUAUACCUCAUAUGUGAA-AUGUGAGCAGUUACCAAGAGCUACCCG------------ ((((..(((....)))((((..((((....((((((-.(((((.....)))))........))))))(((((((.....-)))))))......))))))))))))..------------ ( -26.30, z-score = -2.17, R) >droEre2.scaffold_4770 12109043 105 + 17746568 GGUGCAUGCGAAAGUAGCUCAUUUGGAUAUGUACAC-UUGUUUGUUAAAAAUGUAUCUUUUACAAUCCUCAUAUGCGAA-AUGUGAGCUGUUACCAAGAGCUACCCG------------ ((((..(((....)))((((..((((..........-..............((((.....))))...(((((((.....-)))))))......))))))))))))..------------ ( -24.10, z-score = -1.13, R) >droAna3.scaffold_13340 2586426 101 - 23697760 GGUAGCUGCACAAGUAGCUCAUUUGAAUGUGUGU---UUGGUUAUUUGAGAAGAAAUGCCUAU---UUUUGUACACAAGCUUAUGAGCUGUGUCCCAGGGCUAUCCA------------ ((((((((((((....((((((..(..((((((.---.(((.(((((......))))).))).---.....))))))..)..))))))))))).....)))))))..------------ ( -28.30, z-score = -1.03, R) >consensus GGUGAAUGCGAAAGUAGCUCAUUUGGAUAUGUACAC_UUAUUUGU_AAAAAUGUAUCUUUUGUAUACCUCAUAUGCGAA_AUGUGAGCUGUUACCAAGAGCUACCCG____________ .............(((((((...((((((.........)))))).......................(((((((......)))))))..........)))))))............... (-11.64 = -10.86 + -0.79)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:18 2011