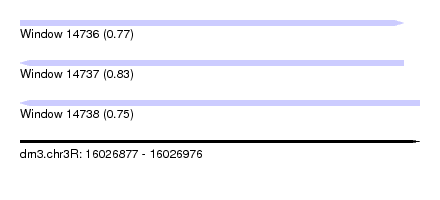
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,026,877 – 16,026,976 |
| Length | 99 |
| Max. P | 0.834424 |

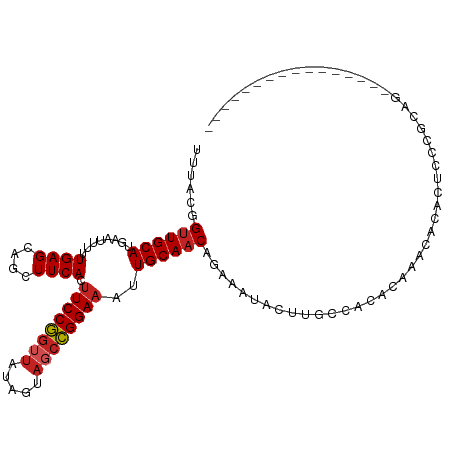
| Location | 16,026,877 – 16,026,972 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Shannon entropy | 0.41822 |
| G+C content | 0.44063 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -15.67 |
| Energy contribution | -15.61 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
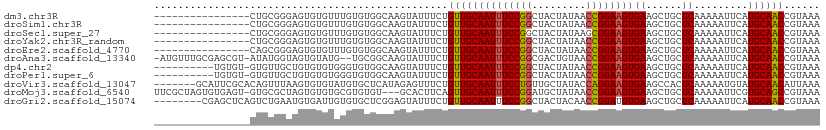
>dm3.chr3R 16026877 95 + 27905053 ----------------CUGCGGGAGUGUGUUUGUGUGGCAAGUAUUUCUGUUGCAAUUUCCGGCUACUAUAACCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA ----------------.(((((..(((((((((.(..((..(((.......))).((((((((.........))))))))...))..).))))....)))))..))))).. ( -30.90, z-score = -2.77, R) >droSim1.chr3R 22061677 95 - 27517382 ----------------CUGCGGGAGUGUGUUUGUGUGGCAAGUAUUUCUGUUGCAAUUUCCGGCUACUAUAACCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA ----------------.(((((..(((((((((.(..((..(((.......))).((((((((.........))))))))...))..).))))....)))))..))))).. ( -30.90, z-score = -2.77, R) >droSec1.super_27 94038 95 - 799419 ----------------CUGCGGGAGUGUGUUUGUGUGGCAAGUAUUUCUGUUGCAAUUUCCGGCUACUAUAAGCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA ----------------.(((((..(((((((((.(..((..(((.......))).(((((((.((......)))))))))...))..).))))....)))))..))))).. ( -29.70, z-score = -2.14, R) >droYak2.chr3R_random 294863 95 + 3277457 ----------------CUGCGGGAGUGUGUUUGUGUGGCAAGUAUUUCUGUUGCAAUUUCCGGCUACUAUAACCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA ----------------.(((((..(((((((((.(..((..(((.......))).((((((((.........))))))))...))..).))))....)))))..))))).. ( -30.90, z-score = -2.77, R) >droEre2.scaffold_4770 12100125 95 - 17746568 ----------------CAGCGGGAGUGUGUUUGUGUGGCAAGUAUUUCUGUUGCAAUUUCCGGCUACUAUAACCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA ----------------..((((..(((((((((.(..((..(((.......))).((((((((.........))))))))...))..).))))....)))))..))))... ( -30.00, z-score = -2.33, R) >droAna3.scaffold_13340 2577622 107 + 23697760 -AUGUUUGCGAGCGU-AUAUGGUAGUGUAUG--UGCGGCAAGUAUUUCUGUUGCAAUUUCCGGCGACUGUAACCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA -(((.(((((.((((-((((....)))))))--)(((((..(((.......))).((((((((.........))))))))...)))))...........))))).)))... ( -30.70, z-score = -0.98, R) >dp4.chr2 3131831 100 - 30794189 ----------UGUGU-GUGUUGCUGUGUGUGGGUGUGGCAAGUAUUUCUGUUGCAAUUUCCGGCUACUAUAACCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA ----------.....-(.(((((((.((.((((((..(((........)))..).((((((((.........))))))))......)))))...)).)).))))))..... ( -27.20, z-score = -0.94, R) >droPer1.super_6 3155124 100 - 6141320 ----------UGUGU-GUGUUGCUGUGUGUGGGUGUGGCAAGUAUUUCUGUUGCAAUUUCCGGCUACUAUAACCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA ----------.....-(.(((((((.((.((((((..(((........)))..).((((((((.........))))))))......)))))...)).)).))))))..... ( -27.20, z-score = -0.94, R) >droVir3.scaffold_13047 2866779 104 + 19223366 -------GCAUUCGCACAGUUUAAGUGUGUAUGUGCUCAUAGAGUUUCUGUUGCAAUUUCCUGUUGCUAUACCAGGAAGUGAAGCCACUCAAAAAUGUAUGCAACAUUAAA -------((((.(((((.......)))))...))))............(((((((..((((((.........))))))(((....)))...........)))))))..... ( -26.40, z-score = -1.37, R) >droMoj3.scaffold_6540 14211867 107 - 34148556 UUCGCUAGUGUGAGU-GUGCGCUAGUGUGUGCGUGUGU---GCACUUCAGUUGCAAUUUCCGGAUGCUAUAACCGGAAGUGAAGCUGCUCAAAAAUUCGUGCAGCCGUAAA ..(((((((((....-..))))))))).(((((....)---)))).....((((.((((((((.........))))))))...(((((.(........).))))).)))). ( -36.90, z-score = -2.13, R) >droGri2.scaffold_15074 480603 103 - 7742996 --------CGAGCUCAGUCUGAAUGUGAUUGUGUGCUCGGAGUAUUUCUGUUGCAAUUUCCGGCUACUACAACCGGAUGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA --------(((((.(((((.......)))))...)))))..........(((((....(((((.........))))).(((((..(......)..))))))))))...... ( -27.60, z-score = -1.26, R) >consensus ________________CUGCGGGAGUGUGUGUGUGUGGCAAGUAUUUCUGUUGCAAUUUCCGGCUACUAUAACCGGAAGUGAAGCUGCUCAAAAAUUCAUGCAACCGUAAA .................................................((((((((((((((.........))))))))((......)).........))))))...... (-15.67 = -15.61 + -0.06)

| Location | 16,026,877 – 16,026,972 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Shannon entropy | 0.41822 |
| G+C content | 0.44063 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
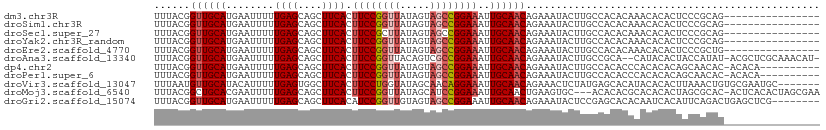
>dm3.chr3R 16026877 95 - 27905053 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG---------------- ......((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).))).))..................---------------- ( -21.00, z-score = -1.43, R) >droSim1.chr3R 22061677 95 + 27517382 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG---------------- ......((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).))).))..................---------------- ( -21.00, z-score = -1.43, R) >droSec1.super_27 94038 95 + 799419 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGCUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG---------------- ......((.(((.(.(((((((.((((......(((((((......)).))))).))))..))))))).).))).))..................---------------- ( -16.60, z-score = -0.22, R) >droYak2.chr3R_random 294863 95 - 3277457 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG---------------- ......((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).))).))..................---------------- ( -21.00, z-score = -1.43, R) >droEre2.scaffold_4770 12100125 95 + 17746568 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCUG---------------- ......((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).))).))..................---------------- ( -21.00, z-score = -1.25, R) >droAna3.scaffold_13340 2577622 107 - 23697760 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUACAGUCGCCGGAAAUUGCAACAGAAAUACUUGCCGCA--CAUACACUACCAUAU-ACGCUCGCAAACAU- ......((((((........((((....)))).(((((((.......)))))))..))))))........((((.((.--.(((........)))-..))..))))....- ( -20.20, z-score = -0.17, R) >dp4.chr2 3131831 100 + 30794189 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACCCACACACAGCAACAC-ACACA---------- .....(((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).)))...)))...............-.....---------- ( -21.40, z-score = -1.79, R) >droPer1.super_6 3155124 100 + 6141320 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACCCACACACAGCAACAC-ACACA---------- .....(((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).)))...)))...............-.....---------- ( -21.40, z-score = -1.79, R) >droVir3.scaffold_13047 2866779 104 - 19223366 UUUAAUGUUGCAUACAUUUUUGAGUGGCUUCACUUCCUGGUAUAGCAACAGGAAAUUGCAACAGAAACUCUAUGAGCACAUACACACUUAAACUGUGCGAAUGC------- .....(((((((...........(((....)))((((((.........))))))..))))))).......(((..(((((.............)))))..))).------- ( -23.12, z-score = -0.92, R) >droMoj3.scaffold_6540 14211867 107 + 34148556 UUUACGGCUGCACGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGCAUCCGGAAAUUGCAACUGAAGUGC---ACACACGCACACACUAGCGCAC-ACUCACACUAGCGAA .....((((((.(((....))).))))))....((((((.........)))))).((((.......((((---......))))....(((.(...-.....).))))))). ( -27.20, z-score = -1.81, R) >droGri2.scaffold_15074 480603 103 + 7742996 UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACAUCCGGUUGUAGUAGCCGGAAAUUGCAACAGAAAUACUCCGAGCACACAAUCACAUUCAGACUGAGCUCG-------- ......((((((........((((....))))..((((((((...))))))))...))))))..........(((((...((.((.......)).)).)))))-------- ( -24.20, z-score = -1.01, R) >consensus UUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG________________ ......((((((........((((....)))).((((((((.....))))))))..))))))................................................. (-16.44 = -16.82 + 0.38)

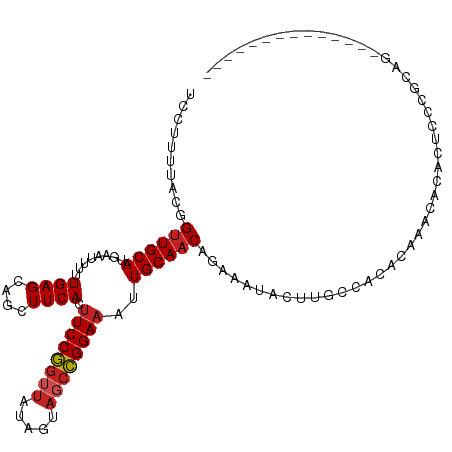
| Location | 16,026,877 – 16,026,976 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Shannon entropy | 0.40915 |
| G+C content | 0.44070 |
| Mean single sequence MFE | -21.39 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16026877 99 - 27905053 UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG-------------- ..........((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).))).))..................-------------- ( -21.00, z-score = -1.29, R) >droSim1.chr3R 22061677 99 + 27517382 UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG-------------- ..........((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).))).))..................-------------- ( -21.00, z-score = -1.29, R) >droSec1.super_27 94038 99 + 799419 UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGCUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG-------------- ..........((.(((.(.(((((((.((((......(((((((......)).))))).))))..))))))).).))).))..................-------------- ( -16.60, z-score = -0.17, R) >droYak2.chr3R_random 294863 99 - 3277457 UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG-------------- ..........((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).))).))..................-------------- ( -21.00, z-score = -1.29, R) >droEre2.scaffold_4770 12100125 99 + 17746568 UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCUG-------------- ..........((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).))).))..................-------------- ( -21.00, z-score = -1.15, R) >droAna3.scaffold_13340 2577622 111 - 23697760 UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUACAGUCGCCGGAAAUUGCAACAGAAAUACUUGCCGCAC--AUACACUACCAUAUACGCUCGCAAACAU ..........((((((........((((....)))).(((((((.......)))))))..))))))........((((.((..--(((........)))..))..)))).... ( -20.20, z-score = -0.13, R) >dp4.chr2 3131832 103 + 30794189 UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACCCACACACAGCAACACACAC---------- .........(((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).)))...)))...................---------- ( -21.40, z-score = -1.68, R) >droPer1.super_6 3155125 103 + 6141320 UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACCCACACACAGCAACACACAC---------- .........(((.(((.(.(((((((.((((......(((((((((...))))))))).))))..))))))).).)))...)))...................---------- ( -21.40, z-score = -1.68, R) >droVir3.scaffold_13047 2866783 104 - 19223366 UUCUUUUAAUGUUGCAUACAUUUUUGAGUGGCUUCACUUCCUGGUAUAGCAACAGGAAAUUGCAACAGAAACUCUAUGAGCACAUACACACUUAAACUGUGCGA--------- .........(((((((...........(((....)))((((((.........))))))..)))))))............(((((.............)))))..--------- ( -22.82, z-score = -1.16, R) >droMoj3.scaffold_6540 14211868 110 + 34148556 UUCUUUUACGGCUGCACGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGCAUCCGGAAAUUGCAACUGAAGUGCA---CACACGCACACACUAGCGCACACUCACACUAGCGA (((......((((((.(((....))).))))))....((((((.........)))))).........)))((((.---.....)))).(.((((.(........).)))).). ( -27.10, z-score = -1.74, R) >droGri2.scaffold_15074 480604 106 + 7742996 UUCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACAUCCGGUUGUAGUAGCCGGAAAUUGCAACAGAAAUACUCCGAGCACACAAUCACAUUCAGACUGAGCUC------- ..........((((((........((((....))))..((((((((...))))))))...))))))...........((((...((.((.......)).)).))))------- ( -21.80, z-score = -0.09, R) >consensus UCCUUUUACGGUUGCAUGAAUUUUUGAGCAGCUUCACUUCCGGUUAUAGUAGCCGGAAAUUGCAACAGAAAUACUUGCCACACAAACACACUCCCGCAG______________ ..........((((((........((((....)))).((((((((.....))))))))..))))))............................................... (-16.44 = -16.82 + 0.38)

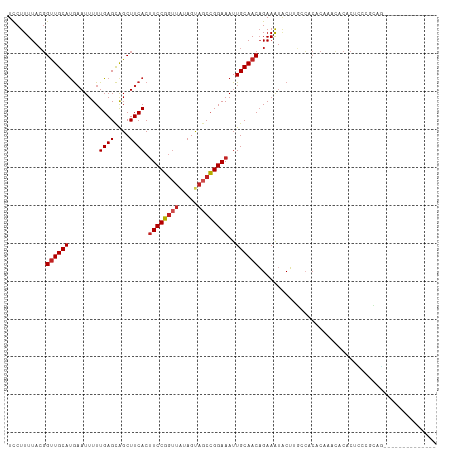
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:16 2011