| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,016,802 – 16,016,946 |
| Length | 144 |
| Max. P | 0.649194 |

| Location | 16,016,802 – 16,016,946 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 162 |
| Reading direction | forward |
| Mean pairwise identity | 76.59 |
| Shannon entropy | 0.41047 |
| G+C content | 0.33513 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -19.38 |
| Energy contribution | -20.06 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
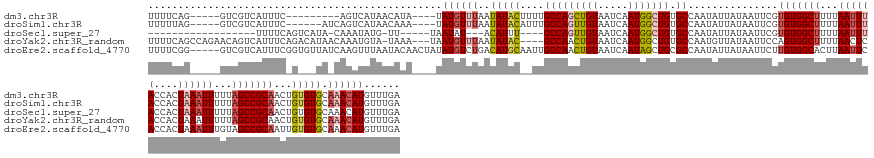
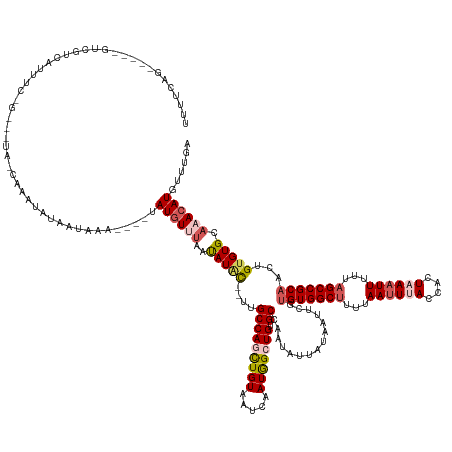
>dm3.chr3R 16016802 144 + 27905053 UUUUCAG-----GUCGUCAUUUC---------AGUCAUAACAUA----UAUGUUUAAUAUACUUUUGCCAGCUGUAAUCAAUGGCUGUGCCAAUAUUAUAAUUCGUGUGGCUUUUAAUUUACCACUAAAUUUUUAGCCGCAACUGUGUGCAAACAUGUUUGA .......-----...........---------........((.(----(((((((..(((((....(((((((((.....))))))).))...............(((((((...((((((....))))))...)))))))...))))).)))))))).)). ( -32.60, z-score = -1.71, R) >droSim1.chr3R 22051567 147 - 27517382 UUUUUAG-----GUCGUCAUUUC------AUCAGUCAUAACAAA----UAUGUUUAAUAUACAUUUGCCAGUUGUAAUCAAUGGCUGUGCCAAUAUUAUAAUUCGUGUGGCUUUUAAUUUACCACUAAAUUUUUAGCCGCAACUGUGUGCAAACAUGUUUGA .......-----...........------...........((((----(((((((..((((((...((.(((((((((...((((...))))..))))))))).))((((((...((((((....))))))...))))))...)))))).))))))))))). ( -36.50, z-score = -2.86, R) >droSec1.super_27 83636 130 - 799419 ------------------UUUUCAGUCAUA-CAAAUAUG-UU-----UAAUAU---ACAUUU----GCCAGUUGUAAUCAAUGGCUGUGCCAAUAUUAUAAUUCGUGUGGCUUUUAAUUUACCACUAAAUUUUUAGCCGCAACUGUGUGCAAACAUGUUUGA ------------------............-((((((((-((-----(..(((---(((...----((.(((((((((...((((...))))..))))))))).))((((((...((((((....))))))...))))))...)))))).))))))))))). ( -36.50, z-score = -3.82, R) >droYak2.chr3R_random 2638771 154 + 3277457 UUUUCAGCCAGAACAGUCAUUUCAGACAUAACAAAUGUA-UAAA---UAAUGUUUAAUAUAC----GCCAACUGUAAUCAAUGGCUGUGCCAAUGUUAUAAUUCCAGUGGCUUUUAACUCACCACUAAAUUUUUAGCCGCAACUGUGUGCAAACAUGUUUGA ....(((((((((((...((((...((((.....)))).-.)))---)..)))))....(((----(.....)))).....)))))).....(((((........(((((...........))))).........(((((....))).)).)))))...... ( -27.10, z-score = 0.53, R) >droEre2.scaffold_4770 12089238 157 - 17746568 UUUUCGG-----GUCGUCAUUUCGGUGUUAUCAAGUUUAAUACAACUAUAUGUCUGACAUGCAAUUGCCAACUGUAAUCAAUAGCUGCGCCAAUAUUAUAAUUCUUGUGGCACUUAAUUCACCACUAAAUUUGUAGCCGCAAUUGUGUGCAAACAUGUUUGA ....(((-----(.......))))((((((.......))))))....((((((.((.(((((((((((....((....))...(((((((((...............))))....((((........)))).))))).))))))))))))).)))))).... ( -31.96, z-score = 0.28, R) >consensus UUUUCAG_____GUCGUCAUUUC_G___UA_CAAAUAUAAUAAA____UAUGUUUAAUAUAC__UUGCCAGCUGUAAUCAAUGGCUGUGCCAAUAUUAUAAUUCGUGUGGCUUUUAAUUUACCACUAAAUUUUUAGCCGCAACUGUGUGCAAACAUGUUUGA .........................................................(((((....(((((((((.....))))))).))...............(((((((...((((((....))))))...)))))))...)))))((((....)))). (-19.38 = -20.06 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:13 2011