
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,010,331 – 16,010,436 |
| Length | 105 |
| Max. P | 0.599902 |

| Location | 16,010,331 – 16,010,436 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.39763 |
| G+C content | 0.46601 |
| Mean single sequence MFE | -31.61 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.14 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
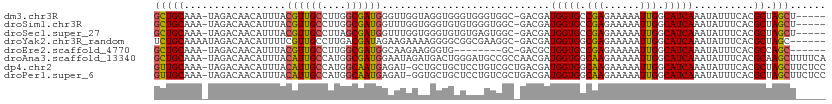
>dm3.chr3R 16010331 105 + 27905053 GCUGCAAA-UAGACAACAUUUACGUUGCCUUGGCGAUGGGUUGGUAGGUGGGUGGGUGGC-GACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCU----- (((((...-....((((.....((((((....)))))).))))....((((((.(....)-.))..(((((((((......)))))))))......)))))).))).----- ( -34.10, z-score = -1.74, R) >droSim1.chr3R 22045097 105 - 27517382 GCUGCAAA-UAGACAACAUUUACGUUGCCUUGGCGAUGGUUUGGUGGGUGUGUGGGUGGC-GACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCU----- (((((...-((.(((.((((..((((((....))))))....))))..))).)).((((.-.....(((((((((......)))))))))......)))))).))).----- ( -34.80, z-score = -1.76, R) >droSec1.super_27 77375 105 - 799419 GCUGCAAA-UAGACAACAUUUACGUUGCCUUAGCGAUGGUUUGGUGGGUGUGUGAGUGGC-GACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCU----- (((((...-......((((((((...(((........)))...))))))))(((((((..-..)).(((((((((......))))))))).....))))))).))).----- ( -34.10, z-score = -1.90, R) >droYak2.chr3R_random 276774 105 + 3277457 UCUGCAAAAUAGACAACAUUUUCGUUGCCUUGACGAUAGAAGAAAAGGGGCGGCGAAGGC-GACGAUGGUGGCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGC------ ...((...........(((..((((((((((..((...............))...)))))-)))))..)))(((.((((.(((.......))).)))).)))..))------ ( -26.16, z-score = -1.02, R) >droEre2.scaffold_4770 12082634 96 - 17746568 GCUGCAAA-UAGACAACAUUUACGUUGCCUUGGCGAUGGCAAGAAGGGUG--------GC-GACGCUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCCAGC------ (((((...-.............(((..((((.((....))....))))..--------))-)....(((((((((......)))))))))..........).))))------ ( -31.20, z-score = -1.72, R) >droAna3.scaffold_13340 2561929 111 + 23697760 GCUGCAAA-UAGACAACAUUUACAUUGCCAUGGCGAUGGAAUAGAUGACUGGGAUGCCGCCAACGAUGGUGGCAAGAAAAAUUGGCAUCAAAUAUUUCACGCAAGCUUUUCA (((((...-.............((((((....)))))).....((((.(..(..((((((((....)))))))).......)..)))))...........)).)))...... ( -32.40, z-score = -2.18, R) >dp4.chr2 3116244 110 - 30794189 GUUGCAAA-UAGACAACAUUUACAUUGCCAUGGCAAUGAGAU-GCUGCUGCUCCUGUCGCUGACGAUGGUGGCAAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCUUCUCC ((((....-....)))).....((((((....))))))(((.-((((((((..((((((....))))))..))).((((.(((.......))).))))..)).))).))).. ( -31.30, z-score = -1.43, R) >droPer1.super_6 3139614 110 - 6141320 GUUGCAAA-UAGACAACAUUUACAUUGCCAUGGCAAUGAGAU-GGUGCUGCUCCUGUCGCUGACGAUGGUGGCAAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCUUCUCC (((((...-.((.((.(((((.((((((....))))))))))-).))))((..((((((....))))))..))..((((.(((.......))).))))..)).)))...... ( -28.80, z-score = -0.52, R) >consensus GCUGCAAA_UAGACAACAUUUACGUUGCCUUGGCGAUGGGAUGGAGGGUGCGUCGGUGGC_GACGAUGGUGCCGAGAAAAAUUGGCAUCAAAUAUUUCACGCUAGCU_____ (((((.................((((((....))))))............................(((((.(((......))).)))))..........)).)))...... (-18.60 = -18.14 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:12 2011