
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,970,431 – 15,970,618 |
| Length | 187 |
| Max. P | 0.646711 |

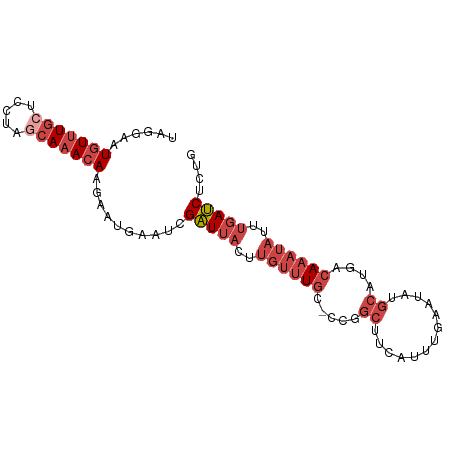
| Location | 15,970,431 – 15,970,521 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 76.36 |
| Shannon entropy | 0.46309 |
| G+C content | 0.34900 |
| Mean single sequence MFE | -18.23 |
| Consensus MFE | -9.60 |
| Energy contribution | -10.35 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15970431 90 + 27905053 UAGGAAUGUUUGCUCCUAGCAAACAAGAAUGAAUCGAUUACUUGUUUGC-CCGGCUUCAUUUGAAUAUGCAUGACAAAUAUUUGAUCUCUG ...(((((((((.((...((((((((((((......))).)))))))))-...(((((....)))...))..)))))))))))........ ( -19.60, z-score = -1.67, R) >droPer1.super_6 3110535 86 - 6141320 UAUGGAUUUAUAAUCUCUGCAAACAUAAAGAUAUAGGUAUAUAAUUUAUUCUUUCCUUUCAUGCAAAUAUAUG-CAUUGCCCCCACC---- ...((((.....))))..((((....(((((.((((((.....))))))))))).......((((......))-)))))).......---- ( -9.20, z-score = 0.64, R) >droEre2.scaffold_4770 12051243 91 - 17746568 UAGGAGUGUUUGCUCCAAGCAAACAAAAAUGAAUCGAUUACUUGUUUGCACCGGCUCCAUUCGAAUAUGCAAGACAAAUAUUUGAUCUCCG ..(((((((((((.....))))))).......(((((.((.(((((((((.(((......)))....))).)))))).)).))))))))). ( -24.10, z-score = -3.03, R) >droYak2.chr3R 4780012 91 + 28832112 UAGAAAUGUUUGCUCCAAGCAAACAACAAUGAAUCGAUUACUUGUUUGCCACGGCUUCAUUUGAAUAUGCAUGACAAAUAUUUGAUCUCUG ((((..(((((((.....)))))))..........(((((..((((((.((..(((((....)))...)).)).))))))..))))))))) ( -19.80, z-score = -2.04, R) >droSec1.super_27 41837 90 - 799419 UAGGAAUGUUUGCUUAUAUCAAACAAGAAUGAAUCGAUUACUUGUUUGC-CCGGCUUCAUUUGAAUAUGCAUGACAAAUAUUUGAUCUCUG ...(((((((((.((((..(((((((((((......))).)))))))).-...(((((....)))...)))))))))))))))........ ( -16.40, z-score = -0.82, R) >droSim1.chr3R 22013695 90 - 27517382 UAGGAAUGUUUGCUUAUAGCAAACAAGAAUGAAUCGAUUACUUGUUUGC-CCGGCUUCAUUUGAAUAUGCAUGACAAAUAUUUGAUCUCUG ...(((((((((.((((.((((((((((((......))).)))))))))-...(((((....)))...)))))))))))))))........ ( -20.30, z-score = -1.98, R) >consensus UAGGAAUGUUUGCUCCUAGCAAACAAGAAUGAAUCGAUUACUUGUUUGC_CCGGCUUCAUUUGAAUAUGCAUGACAAAUAUUUGAUCUCUG ......(((((((.....))))))).......(((((.((.((((((((.....(.......).....))).))))).)).)))))..... ( -9.60 = -10.35 + 0.75)

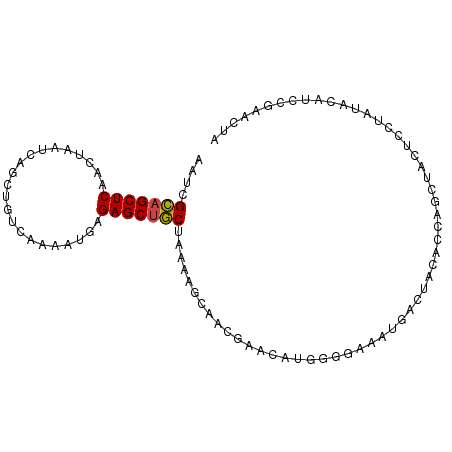
| Location | 15,970,521 – 15,970,618 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.48 |
| Shannon entropy | 0.50704 |
| G+C content | 0.43079 |
| Mean single sequence MFE | -16.68 |
| Consensus MFE | -10.39 |
| Energy contribution | -10.28 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15970521 97 + 27905053 AAUCGCAGCUCAACUAAUCAGCUGUCAAAAUGAGAGCUGCUAAAAGCAACGAACAUGGGUAAAUGACUACACCAGCUACUCCUAUACAUCCGAACUA ..((((((((.........))))))......(((((((((.....))).......((((((......))).)))))).)))..........)).... ( -18.50, z-score = -1.19, R) >droAna3.scaffold_13340 2535053 97 + 23697760 AAUCGACGCUCAACUCAUCAGCUGUCAAAACAAGAGCUUCUGAUACGACAGGAAGCCCGCGAAAGGGGAGGAAUAUAACUAGCUUGCAACUAAACAG ....((((((.........))).)))....((((.((((((.........))))))((.(....).))..............))))........... ( -19.40, z-score = -0.23, R) >droEre2.scaffold_4770 12051334 77 - 17746568 AAUCGCAGCUCAACUAAUCAUCUGUCAAAAUGAGAGCUGCUAAA-GCACAGAACA-------------------GCCACUCCUACACAUCCGAACUA ..(((((((((......((((........)))))))))))....-((........-------------------))...............)).... ( -13.00, z-score = -1.32, R) >droYak2.chr3R 4780103 95 + 28832112 AAUCGCAGCUCAACUAAUCAUCUGUCAAAAUAAGAGCUGCUAAAAGCCA-GAACA-GGGGAAAUGACUACAGCAGCUACUCCUAUACAUCCAAACUA ....(((((((......................))))))).........-....(-((((...((.(....)))....))))).............. ( -16.15, z-score = -0.46, R) >droSec1.super_27 41927 92 - 799419 AAUCGCAGCUCAACUAAUCAGCUGUCAAAAUGAGAGCUGCUAAAAGCAACGAACAUGGGGAAAUGACUACACCUACA-----UAUACAUCCGAACUA ..(((..(((........(((((.((.....)).))))).....)))..)))...((((............))))..-----............... ( -15.92, z-score = -0.94, R) >droSim1.chr3R 22013785 97 - 27517382 AAUCGCAGCUCAACUAAUCAGCUGUCAAAAUGAGAGCUGCUAAAAGCAACGAACAUGGGGAAAUGACUACACCAGCUACUCCUAUACAUCCGAACUA ..(((..(((........(((((.((.....)).))))).....)))..)))..((((((...((.......)).....))))))............ ( -17.12, z-score = -0.54, R) >consensus AAUCGCAGCUCAACUAAUCAGCUGUCAAAAUGAGAGCUGCUAAAAGCAACGAACAUGGGGAAAUGACUACACCAGCUACUCCUAUACAUCCGAACUA ....(((((((......................)))))))......................................................... (-10.39 = -10.28 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:07 2011