
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,075,532 – 8,075,625 |
| Length | 93 |
| Max. P | 0.553234 |

| Location | 8,075,532 – 8,075,625 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 71.07 |
| Shannon entropy | 0.60599 |
| G+C content | 0.49491 |
| Mean single sequence MFE | -27.19 |
| Consensus MFE | -14.55 |
| Energy contribution | -14.52 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8075532 93 - 23011544 -------------UAAUAAGUUUACAAAUUUCAUAGUUGGAUAAACCAAAACGAGUCGGAGCGCUCAGGCAUGGCGGUCUUGGAUGUGGCCAUACCGACUGGCUAU -------------...................((((((((.....))))..(.((((((...((....))(((((.(.(......)).))))).)))))).))))) ( -22.40, z-score = -0.27, R) >droEre2.scaffold_4929 16981143 96 - 26641161 ---------AUUGUAUUGAGUUUGUAAAUUUC-UAGUUGGAUAAACCAAAACGAGUCGGAGCGCUCAGGCAUGGCGGUUUUGGAUGUGGCCAUACCGACUGGCUAU ---------......................(-(((((((.....(((((((..((((..((......)).)))).)))))))(((....))).)))))))).... ( -24.40, z-score = -0.21, R) >droYak2.chr2L 10705583 92 - 22324452 -------------UGUUAAGUUUGUAAAUUUC-UAGUUGGAUAAACCAGAACGAGUCGGAGCGCUCAGGCAUGGCAGUUUUGGAUGUGGCCAUACCGACUGGCUAU -------------..................(-(((((((.....(((((((..((((..((......)).)))).)))))))(((....))).)))))))).... ( -24.30, z-score = -0.49, R) >droSec1.super_3 3572423 93 - 7220098 -------------UAAUAAAUUUACCCAUUUCUUAGUUGGAUAAACCAAAACGAGUCGGAGCGCUCAGGCAUGGCGGUCUUGGAUGUGGCCAUACCGACUGGCUAU -------------.......((((.(((.........))).))))......(.((((((...((....))(((((.(.(......)).))))).)))))).).... ( -22.00, z-score = 0.19, R) >droSim1.chr2L 7856287 93 - 22036055 -------------UAAUAACUUUACAAAUUUCUUAGUUGGAUAAACCAAAACGAGUCGGAGCGCUCAGGCAUGGCGGUCUUGGAUGUGGCCAUACCGACUGGCUAU -------------.......................((((.....))))..(.((((((...((....))(((((.(.(......)).))))).)))))).).... ( -21.90, z-score = -0.21, R) >dp4.chr4_group4 5587693 92 + 6586962 -------------UUAUGGCCUUCUCUUUUUG-CAGCUGGAUAAACACCGCAGAAUCGGAACGCUCGGGCAUGGCCAUAUUGGAUGUGGCCAUACCCACUGGCUAU -------------..((((((((((..(((((-(.(.((......))).))))))..)))).....(((.((((((((((...)))))))))).)))...)))))) ( -36.50, z-score = -3.47, R) >droPer1.super_46 58271 92 - 590583 -------------UUAUGGCCUUCUCUUUUUG-CAGCUGGAUAAACACCGCAGAAUCGGAACGCUCGGGCAUGGCCAUAUUGGAUGUGGCUAUACCCACUGGCUAU -------------..((((((((((..(((((-(.(.((......))).))))))..)))).....(((.((((((((((...)))))))))).)))...)))))) ( -34.20, z-score = -3.07, R) >droAna3.scaffold_12916 11798961 106 + 16180835 UAGCUCUUUCUUAAACUUCUUUUAUCCAAAACUUAGCUGGAUAAAUACCAAUGAGUCGGAGCGCUCUGGCAUGGCUGUGUUGGACGUGGCCAUACCCACGGGCUAC ..(((((..((((.......((((((((.........))))))))......))))..)))))(((((((.(((((..((.....))..)))))..))).))))... ( -37.22, z-score = -3.71, R) >droWil1.scaffold_180708 6318899 88 - 12563649 -------------UAAUGAAUUUUCCAU-----UAGAUGGCUGCACACCGAAGAGUCUGAGCGUUCGGGCAUGGCUGUCCUCGAUGUGGCUAUACCUACUGGCUAU -------------...........(((.-----((((((((..((...(((...(((((......)))))..((....))))).))..)))))..))).))).... ( -21.10, z-score = 0.68, R) >droVir3.scaffold_12963 16272767 96 - 20206255 ---------UAAUCGCUCUCUUUGCCCCCUUG-CAGCUGGGUGAACUUGCAGGAAUCGGAGCGUUCGGGCAUGGCUGUGCUGGAUGUGGCCAUACCCACGGGCUAU ---------.....((((.....((((.((((-((....((....))))))))....)).))....(((.(((((..(.......)..))))).)))..))))... ( -30.90, z-score = 1.09, R) >droMoj3.scaffold_6500 21214208 91 - 32352404 --------------AAUCUGUAUGUAUCAUUC-CAGCUGGGUUAACACCAAGGAAUCGGAGCGUUCGGGCAUGGCUGUGCUAGAUGUGGCCAUACCCACGGGCUAC --------------..(((((((((.((((((-(...(((.......))).))))).)).))))..(((.(((((..(.......)..))))).)))))))).... ( -28.10, z-score = -0.17, R) >droGri2.scaffold_15252 13699259 91 - 17193109 --------------UGGUAAAUCUUUAUUAUG-CAGCUGGGUGAACACCCAAGAAUCGGAACGUUCGGGCAUGGCUGUGCUGGAUGUGGCGAUACCCACCGGCUAC --------------.................(-(((((((((....))))..((((......))))......))))))(((((.((.((.....)))))))))... ( -26.00, z-score = 0.12, R) >anoGam1.chr3R 10368584 92 - 53272125 -------------UGUUUUGUUUUUCCCCCUC-CAGAUGGAUCAACACGCAGGAAUCGAUCCGCUCGGGUAUGGCGGUACUGGAUGUGGCCGUACCGACCGGCUAU -------------..............(((((-(....))).......((.(((.....)))))..)))..(((((((((.((......)))))))).)))..... ( -24.50, z-score = 1.38, R) >consensus _____________UAAUAAGUUUAUCAAUUUC_UAGCUGGAUAAACACCAACGAGUCGGAGCGCUCGGGCAUGGCGGUCUUGGAUGUGGCCAUACCCACUGGCUAU ..................................((((((............((((......)))).((.(((((..(.......)..))))).))..)))))).. (-14.55 = -14.52 + -0.03)

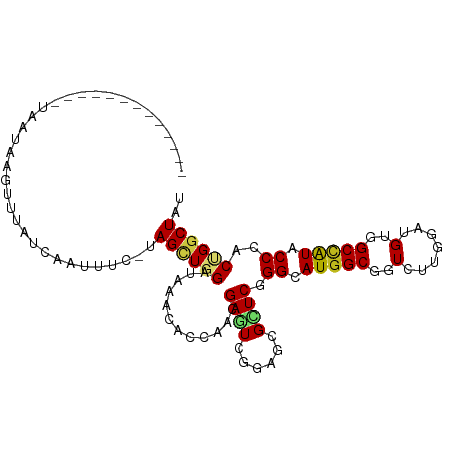

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:01 2011