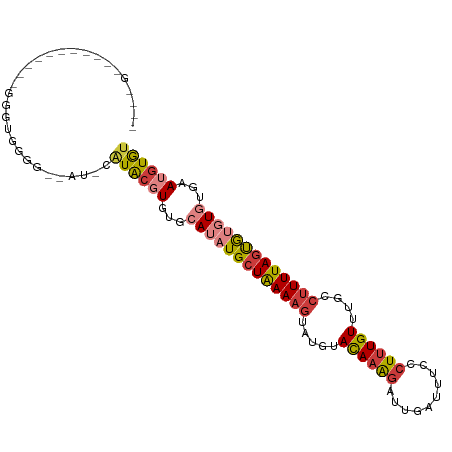
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,909,204 – 15,909,303 |
| Length | 99 |
| Max. P | 0.999090 |

| Location | 15,909,204 – 15,909,303 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 71.27 |
| Shannon entropy | 0.55853 |
| G+C content | 0.39994 |
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.97 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999090 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
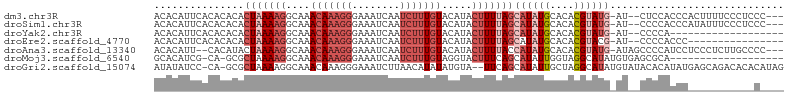
>dm3.chr3R 15909204 99 + 27905053 ---GGGAGGGAAAAGUGGGUGGAG--AU-CAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGUGU ---((((((.(((...(((.((((--((-((....(((((((((.......))))))))).....)))))))))))..))).))))))................. ( -30.20, z-score = -3.64, R) >droSim1.chr3R 21956971 99 - 27517382 ---GGGAGGGAAAUAUGGGUGGGG--AU-CAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGUGU ---((((((.(((((.(((.((..--((-((....(((((((((.......))))))))).....))))..)))))))))).))))))................. ( -30.50, z-score = -3.25, R) >droYak2.chr3R 4695603 83 + 28832112 -------------------UGGGG--AU-CAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGUGU -------------------.....--..-.((((((.(((((((((((((((.....((((((..........))))))....))))))))))))))).)))))) ( -24.30, z-score = -3.26, R) >droEre2.scaffold_4770 11995730 86 - 17746568 ----------------GGGUGGGG--AU-CGUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGUGU ----------------........--..-..(((((.(((((((((((((((.....((((((..........))))))....))))))))))))))).))))). ( -23.40, z-score = -2.29, R) >droAna3.scaffold_13340 2479292 99 + 23697760 ---GGGGCAAGAGGGAGGAUGGGGCUAU-CAUACGUGUGCAUAUGGUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUAUGUG--AAUGUGU ---...((((((((.(((..((((..((-((....(((((((((.......))))))))).....))))..))))...))).)))))).)).....--....... ( -29.10, z-score = -2.23, R) >droMoj3.scaffold_6540 14093949 84 - 34148556 -------------------UGCGCUCACAUAUGCCUACCAAUAUGCUGAAAGUACCUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGCGC-UG-CGAUGUGC -------------------.((((((.(((((........))))((((((((.....((((((..........))))))....))))))))..-.)-.)).)))) ( -19.00, z-score = -1.91, R) >droGri2.scaffold_15074 355895 101 - 7742996 CUAUGUGUGUCUGCUCAUAUGUGUAUACAUAUGCCUAGCAAUAUGCUGAA--UACAUAUAUGUUAAGAUUUCCCUUUGUUUGCCUUUUAGCGC-UG-GGAUAUAU ..((((((((((...(((((((((((.((...((.((....)).)))).)--))))))))))...))))..(((..((((........)))).-.)-)))))))) ( -25.60, z-score = -1.66, R) >consensus ____G___________GGGUGGGG__AU_CAUACGUGUGCAUAUGCUAAAAGUAUGUACAAAGAUUGAUUUCCCUUUGUUUGCCUUUUAGUGUGUGUGAAUGUGU ..............................((((((...(((((((((((((.....((((((..........))))))....)))))))))))))...)))))) (-16.02 = -16.97 + 0.95)


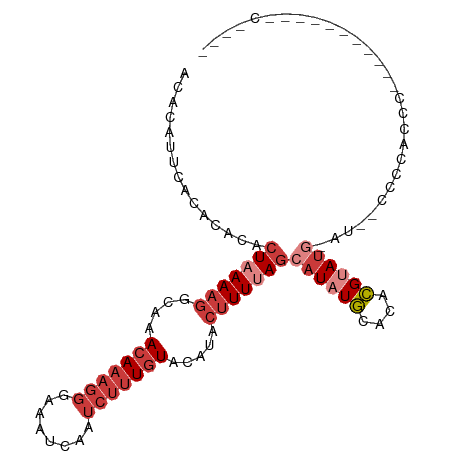
| Location | 15,909,204 – 15,909,303 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.27 |
| Shannon entropy | 0.55853 |
| G+C content | 0.39994 |
| Mean single sequence MFE | -16.33 |
| Consensus MFE | -7.22 |
| Energy contribution | -8.96 |
| Covariance contribution | 1.74 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934233 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 15909204 99 - 27905053 ACACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUAUG-AU--CUCCACCCACUUUUCCCUCCC--- ...............(((((((....(((((((........))))))).....)))))))((((((....))))))-..--.....................--- ( -15.70, z-score = -2.54, R) >droSim1.chr3R 21956971 99 + 27517382 ACACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUAUG-AU--CCCCACCCAUAUUUCCCUCCC--- ...............(((((((....(((((((........))))))).....)))))))((((((....))))))-..--.....................--- ( -15.70, z-score = -2.35, R) >droYak2.chr3R 4695603 83 - 28832112 ACACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUAUG-AU--CCCCA------------------- ...............(((((((....(((((((........))))))).....)))))))((((((....))))))-..--.....------------------- ( -15.70, z-score = -3.04, R) >droEre2.scaffold_4770 11995730 86 + 17746568 ACACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUACG-AU--CCCCACCC---------------- ...............(((((((....(((((((........))))))).....)))))))................-..--........---------------- ( -11.60, z-score = -1.46, R) >droAna3.scaffold_13340 2479292 99 - 23697760 ACACAUU--CACAUACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUACCAUAUGCACACGUAUG-AUAGCCCCAUCCUCCCUCUUGCCCC--- .......--............((((((((((((........)))))))............((((((....))))))-..................)))))..--- ( -16.20, z-score = -1.85, R) >droMoj3.scaffold_6540 14093949 84 + 34148556 GCACAUCG-CA-GCGCUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUAGGUACUUUCAGCAUAUUGGUAGGCAUAUGUGAGCGCA------------------- ((.....)-).-(((((.((((((..(((((((........)))))))..)).))))..((((((........)))))).))))).------------------- ( -21.30, z-score = -1.39, R) >droGri2.scaffold_15074 355895 101 + 7742996 AUAUAUCC-CA-GCGCUAAAAGGCAAACAAAGGGAAAUCUUAACAUAUAUGUA--UUCAGCAUAUUGCUAGGCAUAUGUAUACACAUAUGAGCAGACACACAUAG .....(((-(.-..(((....))).......))))..(((...(((((.((((--(...((((((.((...))))))))))))).)))))...)))......... ( -18.10, z-score = -0.92, R) >consensus ACACAUUCACACACACUAAAAGGCAAACAAAGGGAAAUCAAUCUUUGUACAUACUUUUAGCAUAUGCACACGUAUG_AU__CCCCACCC___________C____ ...............(((((((....(((((((........))))))).....)))))))((((((....))))))............................. ( -7.22 = -8.96 + 1.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:00 2011