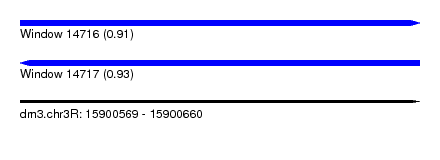
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,900,569 – 15,900,660 |
| Length | 91 |
| Max. P | 0.934738 |

| Location | 15,900,569 – 15,900,660 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.46356 |
| G+C content | 0.42752 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -13.85 |
| Energy contribution | -14.41 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
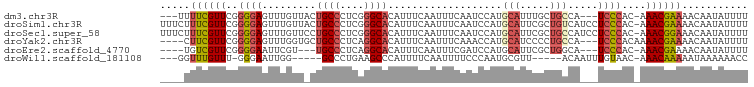
>dm3.chr3R 15900569 91 + 27905053 AAAAUAUUGUUUUCGUUU-GUGGGA---UGGCAGCAAAUGCAUGGAUUGAAAUUGAAAUGUGCCCGAGGGCAGUAACAAACUCCCCGAACGAAAA--- .........(((((((((-(.((((---..(((.....)))...................((((....)))).........))))))))))))))--- ( -26.40, z-score = -2.01, R) >droSim1.chr3R 21948281 97 - 27517382 AAAAUAUUGUUUUCGUUU-GUGGGAGGAUGACAGCGAAUGCAUGGAUUGAAAUUGAAAUGUGCCCGAGGGCAGUAACAAACUCCCCGAACGAAAGAAA .........(((((((((-(.(((((..(((((.(((...((.....))...)))...)))(((....))).....))..)))))))))))))))... ( -31.20, z-score = -3.81, R) >droSec1.super_58 143732 97 - 188520 AAAAUAUUGUUUCCGUUU-GUGGGAGGAUGGCAGCGAAUGCAUGGAUUGAAAUUGAAAUGUGCCCGAGGGCAGGAACAAACUCCCCGAACGAAAGAAA ......((.((((.((((-(.(((((..(((((.....))).................(.((((....)))).)..))..)))))))))))))).)). ( -30.10, z-score = -2.53, R) >droYak2.chr3R 4686864 91 + 28832112 AAAAUAUUGUUUUCGUUUUGUGGGA---UGGCAGGGGAUGCAUGGUUUGAAAUUGAAAUGUGCCUGAGGGCAGCACCAAACUCCCCGAACGAAG---- ...........(((((((.(.((((---.(.....((.(((...((((.......)))).((((....)))))))))...))))))))))))).---- ( -22.50, z-score = 0.24, R) >droEre2.scaffold_4770 11987054 87 - 17746568 AAAAUAUUGUUUUCGUUU-GUGGGA---UGCCAGCGAAUGCAUGGAUCGAAAUUGAAAUGUGCCUGAGGGCA---ACGAAUUCCCCGAACGACA---- ............((((((-(.((((---((((..((...(((((..(((....)))..))))).))..))))---......)))))))))))..---- ( -25.00, z-score = -1.44, R) >droWil1.scaffold_181108 3906622 83 + 4707319 GGUUUUUUAUUUUUGUUU-GUUACAAAUUGU-----AACGCAUUGGGAAAAUUGAAAAUGGGCUUCAGGGC-----CCAAUUCCC-AAACAAACC--- (((((.............-(((((.....))-----)))...(((((((...(....)((((((....)))-----))).)))))-))..)))))--- ( -24.00, z-score = -2.18, R) >consensus AAAAUAUUGUUUUCGUUU_GUGGGA___UGGCAGCGAAUGCAUGGAUUGAAAUUGAAAUGUGCCCGAGGGCAG_AACAAACUCCCCGAACGAAA____ ..........((((((((...((((...((.........((((..............))))(((....))).....))...)))).)))))))).... (-13.85 = -14.41 + 0.56)

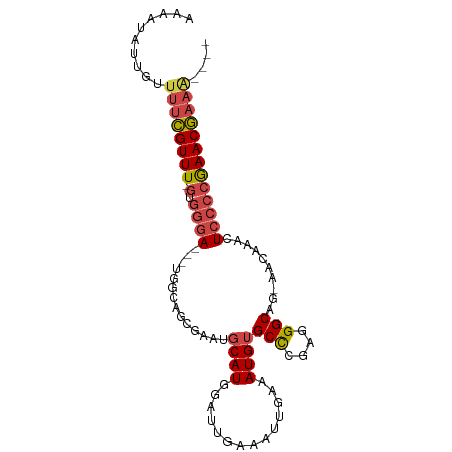
| Location | 15,900,569 – 15,900,660 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.46356 |
| G+C content | 0.42752 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -11.36 |
| Energy contribution | -11.58 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15900569 91 - 27905053 ---UUUUCGUUCGGGGAGUUUGUUACUGCCCUCGGGCACAUUUCAAUUUCAAUCCAUGCAUUUGCUGCCA---UCCCAC-AAACGAAAACAAUAUUUU ---((((((((.(((((..(((....((((....)))).....)))...........(((.....)))..---)))).)-.))))))))......... ( -21.50, z-score = -1.83, R) >droSim1.chr3R 21948281 97 + 27517382 UUUCUUUCGUUCGGGGAGUUUGUUACUGCCCUCGGGCACAUUUCAAUUUCAAUCCAUGCAUUCGCUGUCAUCCUCCCAC-AAACGAAAACAAUAUUUU ....(((((((.((((((..((....((((....))))................((.((....)))).))..))))).)-.))))))).......... ( -24.80, z-score = -3.49, R) >droSec1.super_58 143732 97 + 188520 UUUCUUUCGUUCGGGGAGUUUGUUCCUGCCCUCGGGCACAUUUCAAUUUCAAUCCAUGCAUUCGCUGCCAUCCUCCCAC-AAACGGAAACAAUAUUUU ....(((((((.((((((..((....((((....))))...................(((.....)))))..))))).)-.))))))).......... ( -24.90, z-score = -2.56, R) >droYak2.chr3R 4686864 91 - 28832112 ----CUUCGUUCGGGGAGUUUGGUGCUGCCCUCAGGCACAUUUCAAUUUCAAACCAUGCAUCCCCUGCCA---UCCCACAAAACGAAAACAAUAUUUU ----.((((((.(((((((.((((..((((....))))..............)))).)).)))))((...---.....)).))))))........... ( -20.89, z-score = -1.24, R) >droEre2.scaffold_4770 11987054 87 + 17746568 ----UGUCGUUCGGGGAAUUCGU---UGCCCUCAGGCACAUUUCAAUUUCGAUCCAUGCAUUCGCUGGCA---UCCCAC-AAACGAAAACAAUAUUUU ----..(((((.(((((....((---.(((....)))))..............(((.((....)))))..---)))).)-.)))))............ ( -19.90, z-score = -1.00, R) >droWil1.scaffold_181108 3906622 83 - 4707319 ---GGUUUGUUU-GGGAAUUGG-----GCCCUGAAGCCCAUUUUCAAUUUUCCCAAUGCGUU-----ACAAUUUGUAAC-AAACAAAAAUAAAAAACC ---(((((((((-(((((.(((-----((......))))).........))))))).))(((-----((.....)))))-.............))))) ( -24.10, z-score = -2.69, R) >consensus ____UUUCGUUCGGGGAGUUUGUU_CUGCCCUCAGGCACAUUUCAAUUUCAAUCCAUGCAUUCGCUGCCA___UCCCAC_AAACGAAAACAAUAUUUU .....((((((.((((((.(((....((((....)))).....))).))).......((....)).........)))....))))))........... (-11.36 = -11.58 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:58 2011