
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,884,481 – 15,884,590 |
| Length | 109 |
| Max. P | 0.552965 |

| Location | 15,884,481 – 15,884,590 |
|---|---|
| Length | 109 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 62.19 |
| Shannon entropy | 0.84847 |
| G+C content | 0.38327 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -7.49 |
| Energy contribution | -6.75 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.87 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
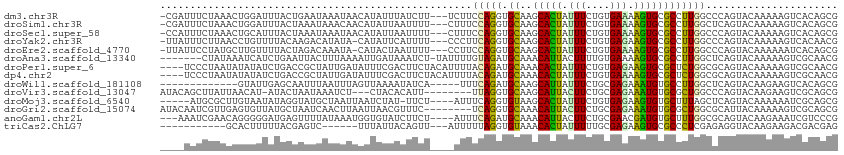
>dm3.chr3R 15884481 109 + 27905053 -CGAUUUCUAAACUGGAUUUACUGAAUAAAUAACAUAUUUAUCUU---UCUUCCAGGUGCAAGCACUAUUUCUGUGAAAAGUGCGCCUUGGCCCAGUACAAAAAGUCACAGCG -.(((((....(((((.........(((((((...)))))))...---....(((((((((..(((.......))).....)))))).))).))))).....)))))...... ( -24.90, z-score = -1.98, R) >droSim1.chr3R 21931247 109 - 27517382 -CGAUUUCUAAACUGGAUUUACUAAAUAAACAACAUAUUAAUUUU---CUUUCCAGGUGCAAGCACUAUUUCUGUGAAAAGUGCGCCUUGGCUCAGUACAAAAAGUCACAGCG -.(((((.....(((((............................---...)))))((((..(((((.((((...)))))))))((....))...))))...)))))...... ( -21.88, z-score = -1.35, R) >droSec1.super_58 9598 109 - 188520 -CCAUUUCUAAACUGCAUUUACUAAAUAAAUAACAUAUUAAUUUU---CUUUCCAGGUGCAAGCACUAUUUCUGUGAAAAGUGCGCCUUGGCCCAGUACAAAAAGUCACAGCG -..........((((.(((((.....)))))..............---....(((((((((..(((.......))).....)))))).)))..))))................ ( -17.30, z-score = -0.26, R) >droYak2.chr3R 4668610 108 + 28832112 -UUAUUUCUUAACCUGUUUUACAAGACAUAUA-CAUAUUCAUUUU---CCCUUCAGGUGCAAGCACUAUUUCUGUGAGAAGUGCGCCUUGGCCCAGUACAAAAAGUCACAACG -.......................(((.....-............---.......((((((..(((.......))).....))))))(((........)))...)))...... ( -16.30, z-score = 0.16, R) >droEre2.scaffold_4770 11969918 108 - 17746568 -UUAUUCCUAUGCUUGUUUUACUAGACAAAUA-CAUACUAAUUUU---CCUUCCAGGUGCAAGCACUAUUUCUGUGAAAAGUGCGCCUUGGCCCAGUACAAAAAAUCACAGCG -..........((((((((....))))))...-............---....(((((((((..(((.......))).....)))))).)))...................)). ( -19.70, z-score = -0.70, R) >droAna3.scaffold_13340 2457283 105 + 23697760 -------CUAUAAAUCAUCUGAAUUACUUUAAAAUUGAUAAAUCU-UAUUUUGUAGAUGCAAACAUUACUUUUGUGAAAAGUGCGCCUUGGCUCAGUACAAAAAGUCGCAACG -------(((((((......((.((((.........).))).)).-...))))))).(((.....((((....))))...((((((....))...))))........)))... ( -12.90, z-score = 0.84, R) >droPer1.super_6 3030965 109 - 6141320 ----UCCCUAAUAUAUAUCUGACCGCUAUUGAUAUUUCGACUUCUACAUUUUACAGAUGCAAACACUAUUUCUGUGAGAAGUGCGCUCUGGCGCAGUACAAAAAGUCGCAACG ----..........(((((...........)))))..((((((.(((.(((((((((.............)))))))))..(((((....))))))))....))))))..... ( -23.62, z-score = -1.60, R) >dp4.chr2 3009647 109 - 30794189 ----UCCCUAAUAUAUAUCUGACCGCUAUUGAUAUUUCGACUUCUACAUUUUACAGAUGCAAACACUAUUUCUGUGAAAAGUGCGCUCUGGCGCAGUACAAAAAGUCGCAACG ----..........(((((...........)))))..((((((.(((.(((((((((.............)))))))))..(((((....))))))))....))))))..... ( -23.52, z-score = -2.08, R) >droWil1.scaffold_181108 784941 95 - 4707319 -------------GUAUUGAGCAAUUUAAUUUAGUUAAAAUAUCA-----UUUCAGAUGCAAGCAUUAUUUCUGCGAGAAAUGUGCCUUGGCUCAGUACAAGAAGUCACAGCG -------------(((((((((..................((((.-----.....))))(((((((.(((((.....)))))))).))))))))))))).............. ( -23.50, z-score = -2.07, R) >droVir3.scaffold_13047 16181925 101 - 19223366 AUACAGCUUAUUAACAU-AUACUAAUAAAUCU---CUACACAUU--------UUAGGUGCAAGCAUUACUUCUGCGAGAAAUGUGCGCUGGCCCAGUACAAGAAGUCGCAGCG ...((((((((((....-....))))))....---...((((((--------((...((((...........)))).)))))))).))))((...(.((.....)))...)). ( -17.30, z-score = 0.98, R) >droMoj3.scaffold_6540 14065461 103 - 34148556 -----AUGCGCUUGUAAUAUAGGUAUGCUAAUUAAUCUAU-UUCU----AUUUCAGGUGUAAGCACUAUUUCUGUGAGAAGUGUGCUUUAGCUCAGUACAAAAAAUCGCAGCG -----.((((((((.(((((((((..........))))).-...)----))).))))))))..........((((((....((((((.......)))))).....)))))).. ( -23.70, z-score = -0.77, R) >droGri2.scaffold_15074 327198 105 - 7742996 AUACAAUCGUUGAGUGUUAUGCUAAUCAACUUAAUUAACGUUUC--------UCAGGUGCAAACAUUACUUCUGCGAGAAAUGUGCGCUGGCGCAUUACAAAAAGUCGCAGCG ........(((((..(.....)...))))).......(((((((--------(((((((.......)))))....))))))))).(((((.((.(((......)))))))))) ( -22.70, z-score = 0.31, R) >anoGam1.chr2L 7200604 106 - 48795086 ---AAAUCGAACAGGGGGAUGAGUUUUAUAAAUGGUGUAUCUUCU----AUUUCAGAUGCAAACAUUACUUCUGCGAACGAUGUGCUUUGGCGCAGUACAAGAAAUCGUCCCG ---............(((((((.((((...((((.(((((((...----.....)))))))..))))....(((((..(((......))).)))))....)))).))))))). ( -28.10, z-score = -1.61, R) >triCas2.ChLG7 9966390 93 + 17478683 -----------GCACUUUUUACGAGUC------UUUAUUACAGUU---AUUUUUAGGUGUAAACACUAUUUUUGCGAGAAGUGCGCCCUCGAGAGGUACAAGAAGACGACGAG -----------((((((((..((((..------....(((((.((---(....))).)))))........))))..))))))))..(((....)))................. ( -20.14, z-score = -0.78, R) >consensus ____UUUCUAAAAUUGAUUUACUAAAUAAAUAAAAUAUUAAUUCU____UUUUCAGGUGCAAGCACUAUUUCUGUGAAAAGUGCGCCUUGGCCCAGUACAAAAAGUCGCAGCG ....................................................(((((.((..(((((.(((....))).))))))))))))...................... ( -7.49 = -6.75 + -0.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:53 2011