| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,806,917 – 15,807,053 |
| Length | 136 |
| Max. P | 0.897377 |

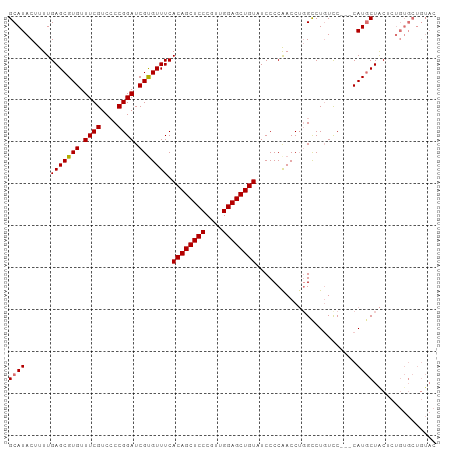
| Location | 15,806,917 – 15,807,016 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.39 |
| Shannon entropy | 0.23501 |
| G+C content | 0.55784 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -22.21 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.828927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15806917 99 + 27905053 GCAUACUUUUGAGCGUGUUUCGGCCCCGGAUCGUGUUUCACAGCUCCCGUUGGAGCUGUUUCCCCAACCAGGUCUGUUC---CAUGCUGCUCUGUGCUGUAC (((((.....(((((.((...((...((((((.((....((((((((....)))))))).....))....))))))..)---)..))))))))))))..... ( -31.70, z-score = -2.04, R) >droSim1.chr3R 21850105 102 - 27517382 GCAUACUUUUGAGCGUGUUUCGUCACCGGAUCGUGUUUCACAGCUCCCGUUGGAGCUGUAUCCCCAACCUGGCCUGUCCCACCAUGCUACUCUGUGCUGUAC (((((......((((((....(.((((((....((....((((((((....)))))))).....))..))))..)).)....))))))....)))))..... ( -28.40, z-score = -1.42, R) >droSec1.super_58 47979 102 - 188520 GCAUACUUUUGAGCGUGUUUCGUCCCCGGAUCGCGUUUCACAGCUCCCGUUGGAGCUGUAUCCCCAACCUGGCCUCUCCCACCAUGCUACUCUGUGCUGUAC (((((.....(((((((.((((....)))).))))))).((((((((....))))))))..........(((......)))...........)))))..... ( -28.40, z-score = -2.09, R) >droEre2.scaffold_4770 11892161 86 - 17746568 GCAUACUUUUGAGCGUGUUUCGUCCCCGGAUCGUGUUUCACAGCUCCCAUUGGAGCUGU-UCCCCGACCUCGCCUGUCC---CAUUCUAC------------ ((........(..((.....))..)..((.(((.(....((((((((....))))))))-...))))))..))......---........------------ ( -19.60, z-score = -1.36, R) >consensus GCAUACUUUUGAGCGUGUUUCGUCCCCGGAUCGUGUUUCACAGCUCCCGUUGGAGCUGUAUCCCCAACCUGGCCUGUCC___CAUGCUACUCUGUGCUGUAC ((((......(((((((.((((....)))).))))))).((((((((....))))))))........................))))............... (-22.21 = -22.27 + 0.06)


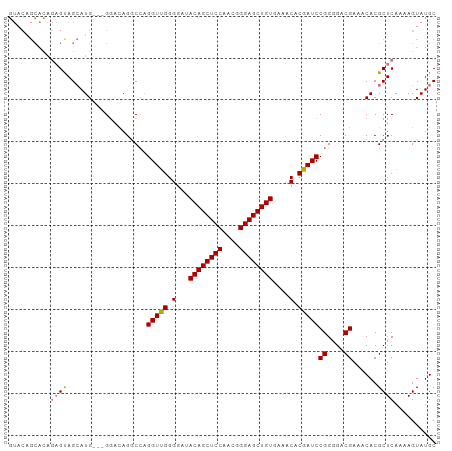
| Location | 15,806,917 – 15,807,016 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.39 |
| Shannon entropy | 0.23501 |
| G+C content | 0.55784 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.897377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15806917 99 - 27905053 GUACAGCACAGAGCAGCAUG---GAACAGACCUGGUUGGGGAAACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGGGCCGAAACACGCUCAAAAGUAUGC ((((.((.....))(((.((---.......(((((((((....((((((((....))))))))....).))).))))).......)).))).....)))).. ( -32.64, z-score = -2.03, R) >droSim1.chr3R 21850105 102 + 27517382 GUACAGCACAGAGUAGCAUGGUGGGACAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGUGACGAAACACGCUCAAAAGUAUGC ((((.((........))...(((......(((.((((((...(((((((((....)))))))))...).))))).))).......)))........)))).. ( -30.02, z-score = -0.80, R) >droSec1.super_58 47979 102 + 188520 GUACAGCACAGAGUAGCAUGGUGGGAGAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACGCGAUCCGGGGACGAAACACGCUCAAAAGUAUGC ...............(((((.(..(((.(....(((((.(..(((((((((....)))))))))...).)))))((....)).....).)))...).))))) ( -33.60, z-score = -2.07, R) >droEre2.scaffold_4770 11892161 86 + 17746568 ------------GUAGAAUG---GGACAGGCGAGGUCGGGGA-ACAGCUCCAAUGGGAGCUGUGAAACACGAUCCGGGGACGAAACACGCUCAAAAGUAUGC ------------........---.....((((.((((((...-((((((((....))))))))....).)))))((....)).....))))........... ( -28.80, z-score = -2.87, R) >consensus GUACAGCACAGAGUAGCAUG___GGACAGGCCAGGUUGGGGAUACAGCUCCAACGGGAGCUGUGAAACACGAUCCGGGGACGAAACACGCUCAAAAGUAUGC ..........((((...................(((((.(...((((((((....))))))))....).)))))((....))......)))).......... (-25.27 = -25.40 + 0.13)

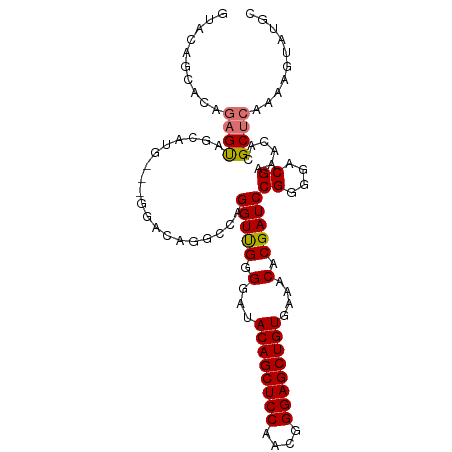
| Location | 15,806,954 – 15,807,053 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.37354 |
| G+C content | 0.47038 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15806954 99 + 27905053 UCACAGCUCCCGUUGGAGCUGUUUCCCCAACCAGGUCUGUUCCA---UGCUGCUCUGUGCUGUACCGCUGCCACAUUAUGUAUUAUAAAAGCAUAAACAUUA ..((((((((....))))))))...........(((..((....---(((.((.....)).)))..)).)))...((((((.........))))))...... ( -21.90, z-score = -0.80, R) >droSim1.chr3R 21850142 102 - 27517382 UCACAGCUCCCGUUGGAGCUGUAUCCCCAACCUGGCCUGUCCCACCAUGCUACUCUGUGCUGUACCACUGCCACAUUAUGUAUUAUAAAAGCAUAAACAUUA ..((((((((....))))))))..........((((.((......)).))))...((((((.......(((........))).......))))))....... ( -22.44, z-score = -1.53, R) >droSec1.super_58 48016 102 - 188520 UCACAGCUCCCGUUGGAGCUGUAUCCCCAACCUGGCCUCUCCCACCAUGCUACUCUGUGCUGUACCACUGCCACAUUAUGUAUUAUAAAAGCAUAAAAAUUA ..((((((((....))))))))..........((((......((.(((........))).)).......))))..((((((.........))))))...... ( -21.12, z-score = -1.87, R) >droYak2.chr3R 4589114 77 + 28832112 UCACUGCUCCCGUUGGAGCUGU-UCCCCGGCCUCGCCUGUCCUA---------------UUC---------CACAUUAUGUAUUAUAAAAGCAUUAACGUUA ..((.(((((....))))).))-.....(((...))).......---------------...---------............................... ( -10.60, z-score = 0.23, R) >droEre2.scaffold_4770 11892198 86 - 17746568 UCACAGCUCCCAUUGGAGCUGU-UCCCCGACCUCGCCUGUCCCA---------------UUCUACCGCUACCACAUUGUGUAUUAUAAAAGCAUAAACGUUG ..((((((((....))))))))-....((((...((..((....---------------....)).)).......((((((.........))))))..)))) ( -16.30, z-score = -2.17, R) >consensus UCACAGCUCCCGUUGGAGCUGU_UCCCCAACCUGGCCUGUCCCA___UGCU_CUCUGUGCUGUACCACUGCCACAUUAUGUAUUAUAAAAGCAUAAACAUUA ..((((((((....)))))))).....................................................((((((.........))))))...... (-13.98 = -14.22 + 0.24)

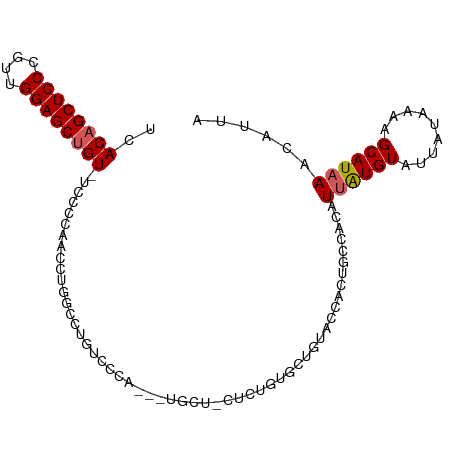

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:50 2011