
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,712,317 – 15,712,413 |
| Length | 96 |
| Max. P | 0.538514 |

| Location | 15,712,317 – 15,712,413 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 102 |
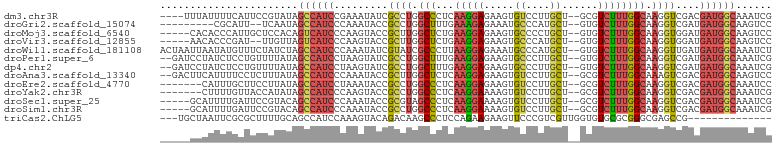
| Reading direction | reverse |
| Mean pairwise identity | 73.14 |
| Shannon entropy | 0.59819 |
| G+C content | 0.50445 |
| Mean single sequence MFE | -25.82 |
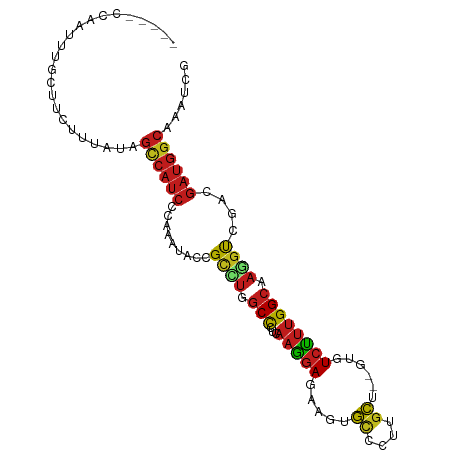
| Consensus MFE | -13.24 |
| Energy contribution | -13.23 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15712317 96 - 27905053 ----UUUAUUUUCAUUCCGUAUAGCCAUCCGAAAUAUCGCCUGGCCCUCAAGGAGAAGUGUCCUUGCU--GCGUCUUUGGCAAGGUCGACGAUGGCAAAUCG ----...................((((((.......((.(((........))).)).(((.(((((((--........))))))).).))))))))...... ( -27.10, z-score = -1.25, R) >droGri2.scaffold_15074 2296500 89 + 7742996 ---------CGCAUU--UCAAUAGCCAUCCCAAAUACCGCCUGGCUUUGAAAGAGAAAUGCCCAUGCU--GUGUCUUUGGCAAGGUCGAUGAUGGCAAGUCC ---------......--......((((((............((((((((..(((((...((....)).--...)))))..))))))))..))))))...... ( -22.44, z-score = -0.24, R) >droMoj3.scaffold_6540 8912568 95 + 34148556 -----CACACCCAUUGCUCCACAGUCAUCCCAAGUACCGCUUGGCUCUGAAGGAGAAGUGCCCCUGCU--GUGUCUUUGGCAAGGUGGAUGAUGGCAAGUCC -----.....((((((.(((((.((((....((((((.((..(((.((........)).)))...)).--))).)))))))...)))))))))))....... ( -26.90, z-score = 0.65, R) >droVir3.scaffold_12855 7414703 93 - 10161210 -----AACACCCGAU--UUGUUAGUCAUCCCAAGUACCGCUUGGCUCUGAAGGAGAAGUGCCCAUGCU--GUGUCUUUGGCAAGGUGGAUGAUGGCAAGUCC -----.......(((--((((((.((((((((((.....))))((..((..(((((...((....)).--...)))))..))..))))))))))))))))). ( -30.20, z-score = -1.39, R) >droWil1.scaffold_181108 605531 100 + 4707319 ACUAAUUAAUAUGUUUCUAUCUAGCCAUCCCAAAUAUCGUAUCGCCCUUAAGGAGAAAUGCCCAUGCU--GUGUCUUUGGCAAGGUUGAUGAUGGCAAAUCU .......................((((((.(((.....((((..(((....)).)..))))((.((((--(......))))).)))))..))))))...... ( -21.30, z-score = -0.59, R) >droPer1.super_6 4511419 98 + 6141320 --GAUCCUAUCUCCUGUUUUAUAGCCAUCCUAAGUAUCGCCUGGCUUUGAAGGAGAAGUGCCCUUGCU--GUGUCUUUGGCAAGGUCGAUGAUGGCAAAUCG --(((.(((((((((.(((((.(((((..(........)..))))).))))).))....(.(((((((--(......)))))))).))).)))))...))). ( -27.50, z-score = -1.16, R) >dp4.chr2 4494163 98 + 30794189 --GAUCCUAUCUCCUGUUUUAUAGCCAUCCUAAGUAUCGCCUGGCUUUGAAGGAGAAGUGCCCUUGCU--GUGUCUUUGGCAAGGUCGAUGAUGGCAAAUCG --(((.(((((((((.(((((.(((((..(........)..))))).))))).))....(.(((((((--(......)))))))).))).)))))...))). ( -27.50, z-score = -1.16, R) >droAna3.scaffold_13340 2293346 98 - 23697760 --GACUUCAUUUUCCUCUUUAUAGCCAUCCCAAAUACCGCUUGGCUCUCAAGGAGAAGUGUCCUUGCU--GCGUCUUUGGCAAAGUCGACGAUGGCAAGUCC --(((....(((((((......(((((..(........)..)))))....)))))))..)))((((((--(((((...(((...))))))).)))))))... ( -26.60, z-score = -1.42, R) >droEre2.scaffold_4770 11795870 93 + 17746568 -------CAUUUGCUUCCUUAUAGCCAUCCUAAAUACCGCCUGGCCCUCAAGGAGAAGUGUCCUUGCU--GCGUCUUUGGCAAGGUCGACGAUGGCAAAUCC -------.((((((((((((...((((..(........)..))))....)))))...(((.(((((((--........))))))).).))...))))))).. ( -26.50, z-score = -1.20, R) >droYak2.chr3R 4483938 93 - 28832112 -------CUUUUGUUACCAUAUAGCCAUCCCAAGUACCGCCUGGCCCUCAAGGAAAAGUGUCCUUGCU--GCGUCUUUGGCAAGGUCGACGAUGGCAAAUCG -------................((((((....(.((((((.((((..((((((......))))))..--).)))...)))..))))...))))))...... ( -27.30, z-score = -1.69, R) >droSec1.super_25 762656 95 + 827797 -----GCAUUUUGAUUCCGUACAGCCAUCCCAAAUACCGCGUAGCCCUCAAGGAAAAGUGUCCUUGCU--GCGUCUUUGGCAAGGUCGACGAUGGCAAAUCG -----..................(((((((((((....(((((((....(((((......))))))))--)))).)))))...(.....)))))))...... ( -26.00, z-score = -1.12, R) >droSim1.chr3R 21755136 95 + 27517382 -----GCAUUUUGAUUCCGUACAGCCAUCCCAAAUACCGCCUGGCCCUCAAGGAAAAGUGUCCUUGCU--GCGUCUUUGGCAAGGUCGACGAUGGCAAAUCG -----..................((((((......((((((.((((..((((((......))))))..--).)))...)))..)))....))))))...... ( -26.10, z-score = -0.81, R) >triCas2.ChLG5 3987508 85 - 18847211 ---UGCUAAUUCGCGCUUUUGCAGCCAUCCAAAGUACAGACAAGCCCUCCAGAAGAAGUUCCCGUCGUUGGUGUGCGCGGGCGAGCCG-------------- ---.(((..(((((((....)).((((.((((......(((..(..((.(....).))..)..))).)))))).)))))))..)))..-------------- ( -20.20, z-score = 1.21, R) >consensus _____CCAAUUUGCUUCUUUAUAGCCAUCCCAAAUACCGCCUGGCCCUCAAGGAGAAGUGCCCUUGCU__GUGUCUUUGGCAAGGUCGACGAUGGCAAAUCG .......................((((((.........((((.(((...((((......((....))......)))).))).))))....))))))...... (-13.24 = -13.23 + -0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:42 2011