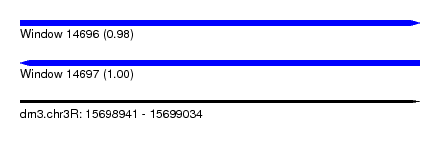
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,698,941 – 15,699,034 |
| Length | 93 |
| Max. P | 0.998497 |

| Location | 15,698,941 – 15,699,034 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 70.85 |
| Shannon entropy | 0.56530 |
| G+C content | 0.43935 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -11.26 |
| Energy contribution | -11.98 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
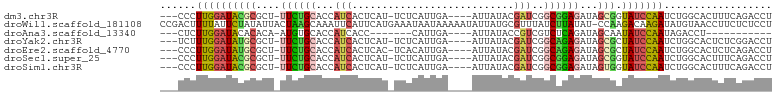
>dm3.chr3R 15698941 93 + 27905053 AGGUCUGAAAGUGCCAGAUUGGAUACCGCUAUCUCCGCCGAUCGUAUAAU----UCAAUGAGA-AUGAGUGAUGGUGCAGAA-AGCGCGUAUCCAAGGG--- .(((.(....).)))...((((((((((((.(((.((((.((((....((----((.....))-))...)))))))).))).-)))).))))))))...--- ( -33.90, z-score = -3.67, R) >droWil1.scaffold_181108 4119292 101 + 4707319 AGGAGAGAAGGUUACAUAUCUUGUCUUGG-AUAUAAGAUAAACGCAUAAUAUUUUUAUUAUUUCAUGAAUGAAUUUGCUUAGUAAUAUAGAAUAAAAGUCGG ..........(((((.....((((((((.-...))))))))..((((((((....))))).((((....))))..)))...)))))................ ( -12.70, z-score = 0.83, R) >droAna3.scaffold_13340 2280995 76 + 23697760 -----------AGGUCUAUUGGAUAUUGCUAUCUGAGACGACGGUAUAAU----UCAAUG-------GGUGAUGGUGCACAU-UGUGUGUAUCCAAGAG--- -----------...((((((((((..(((..((......))..)))..))----))))))-------))...((((((((..-...))))).)))....--- ( -21.40, z-score = -1.65, R) >droYak2.chr3R 4469819 93 + 28832112 AGGUCCGAGAGUGCCAGAUUGGAUAGCGCUAUCUCUGCCGAUCGUAUAAU----UCAAUGAGA-AUGAGUGAUGGUGCAGAA-AGCGCAUAUCCAAAGA--- .((..(....)..))...((((((((((((...((((((.((((....((----((.....))-))...)))).).))))).-))))).)))))))...--- ( -35.60, z-score = -4.40, R) >droEre2.scaffold_4770 11782519 93 - 17746568 AGGUCUGAGAGUGCCAGAUUGGAUAGCGCUAUCUCUGCCGAUCGUAUAAU----UCAAUGUGA-GUGAGUGAUGGUGCAGAA-AGCGCAUAUCCAAGGG--- .(((.(....).)))...((((((((((((...((((((.((((....((----((.....))-))...)))).).))))).-))))).)))))))...--- ( -33.90, z-score = -3.06, R) >droSec1.super_25 749288 93 - 827797 AGGUCUGAAAGUGCCAGAUUGGAUACCGCUAUCUCCGCCGAUCGUAUAAU----UCAAUGAGA-AUGAGUGAUGGUGCAGAA-AGCGCGUAUCCAAGGG--- .(((.(....).)))...((((((((((((.(((.((((.((((....((----((.....))-))...)))))))).))).-)))).))))))))...--- ( -33.90, z-score = -3.67, R) >droSim1.chr3R 21741482 93 - 27517382 AGGUCUGAAAGUGCCAGAUUGGAUACCACUAUCUCCGCCGAUCGUAUAAU----UCAAUGAGA-AUGAGUGAUGGUGCAGAA-AGCGCGUAUCCAAGGG--- .(((.(....).)))...(((((((((.((.(((.((((.((((....((----((.....))-))...)))))))).))).-)).).))))))))...--- ( -28.50, z-score = -2.19, R) >consensus AGGUCUGAAAGUGCCAGAUUGGAUACCGCUAUCUCCGCCGAUCGUAUAAU____UCAAUGAGA_AUGAGUGAUGGUGCAGAA_AGCGCGUAUCCAAGGG___ ..................(((((((((((..(((.((((.((((.........................)))))))).)))...))).))))))))...... (-11.26 = -11.98 + 0.72)


| Location | 15,698,941 – 15,699,034 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 70.85 |
| Shannon entropy | 0.56530 |
| G+C content | 0.43935 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.59 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.998497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15698941 93 - 27905053 ---CCCUUGGAUACGCGCU-UUCUGCACCAUCACUCAU-UCUCAUUGA----AUUAUACGAUCGGCGGAGAUAGCGGUAUCCAAUCUGGCACUUUCAGACCU ---...((((((((.((((-.(((.(.(((((....((-((.....))----)).....))).)).).))).))))))))))))(((((.....)))))... ( -30.60, z-score = -4.12, R) >droWil1.scaffold_181108 4119292 101 - 4707319 CCGACUUUUAUUCUAUAUUACUAAGCAAAUUCAUUCAUGAAAUAAUAAAAAUAUUAUGCGUUUAUCUUAUAU-CCAAGACAAGAUAUGUAACCUUCUCUCCU .................(((((((((...((((....))))((((((....))))))..)))))....((((-(........)))))))))........... ( -7.50, z-score = 0.17, R) >droAna3.scaffold_13340 2280995 76 - 23697760 ---CUCUUGGAUACACACA-AUGUGCACCAUCACC-------CAUUGA----AUUAUACCGUCGUCUCAGAUAGCAAUAUCCAAUAGACCU----------- ---((.(((((((....((-(((.(........).-------))))).----........((..((...))..))..))))))).))....----------- ( -12.40, z-score = -1.32, R) >droYak2.chr3R 4469819 93 - 28832112 ---UCUUUGGAUAUGCGCU-UUCUGCACCAUCACUCAU-UCUCAUUGA----AUUAUACGAUCGGCAGAGAUAGCGCUAUCCAAUCUGGCACUCUCGGACCU ---...(((((((.(((((-((((((.(.(((....((-((.....))----)).....))).)))))))..))))))))))))(((((.....)))))... ( -29.20, z-score = -3.82, R) >droEre2.scaffold_4770 11782519 93 + 17746568 ---CCCUUGGAUAUGCGCU-UUCUGCACCAUCACUCAC-UCACAUUGA----AUUAUACGAUCGGCAGAGAUAGCGCUAUCCAAUCUGGCACUCUCAGACCU ---...(((((((.(((((-((((((.(.(((......-((.....))----.......))).)))))))..))))))))))))(((((.....)))))... ( -27.22, z-score = -3.60, R) >droSec1.super_25 749288 93 + 827797 ---CCCUUGGAUACGCGCU-UUCUGCACCAUCACUCAU-UCUCAUUGA----AUUAUACGAUCGGCGGAGAUAGCGGUAUCCAAUCUGGCACUUUCAGACCU ---...((((((((.((((-.(((.(.(((((....((-((.....))----)).....))).)).).))).))))))))))))(((((.....)))))... ( -30.60, z-score = -4.12, R) >droSim1.chr3R 21741482 93 + 27517382 ---CCCUUGGAUACGCGCU-UUCUGCACCAUCACUCAU-UCUCAUUGA----AUUAUACGAUCGGCGGAGAUAGUGGUAUCCAAUCUGGCACUUUCAGACCU ---...((((((((.((((-.(((.(.(((((....((-((.....))----)).....))).)).).))).))))))))))))(((((.....)))))... ( -28.70, z-score = -3.36, R) >consensus ___CCCUUGGAUACGCGCU_UUCUGCACCAUCACUCAU_UCUCAUUGA____AUUAUACGAUCGGCGGAGAUAGCGGUAUCCAAUCUGGCACUUUCAGACCU ......((((((((((....((((((...(((...........................)))..))))))...))).))))))).................. (-11.44 = -11.59 + 0.15)
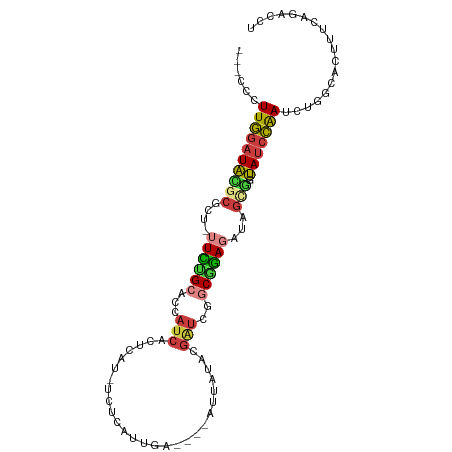
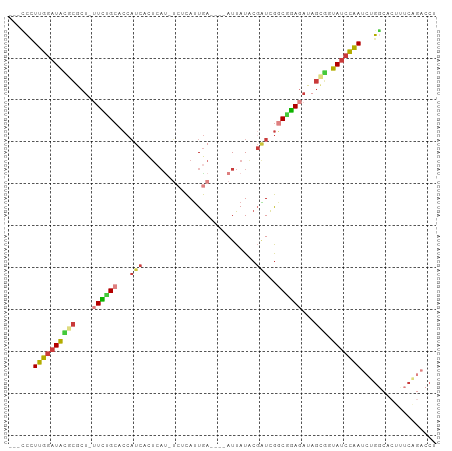
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:41 2011