| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,060,768 – 8,060,874 |
| Length | 106 |
| Max. P | 0.913259 |

| Location | 8,060,768 – 8,060,874 |
|---|---|
| Length | 106 |
| Sequences | 14 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.57 |
| Shannon entropy | 0.61440 |
| G+C content | 0.47145 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -11.66 |
| Energy contribution | -10.16 |
| Covariance contribution | -1.49 |
| Combinations/Pair | 2.38 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
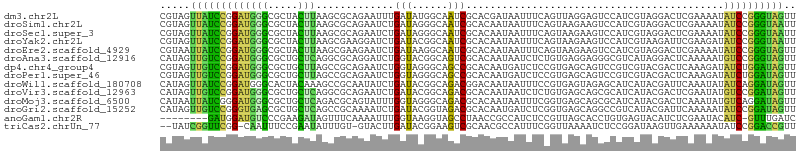
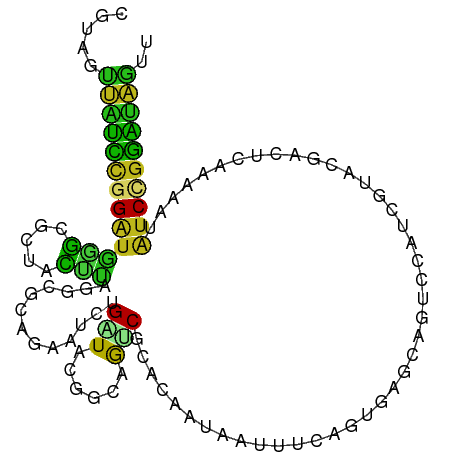
>dm3.chr2L 8060768 106 + 23011544 CGUAGUUAUCCGGAUGGGCGCUACUUAAGCGCAGAAUUUGAUAUGGCAAUCGCACGAUAAUUUCAGUUAGGAGUCCAUCGUAGGACUCGAAAAUAUCCGGGUAGUU .....(((((((((((.(((((.....))))).(((....((...((....))...))...)))......((((((......)))))).....))))))))))).. ( -35.10, z-score = -3.11, R) >droSim1.chr2L 7840191 106 + 22036055 CGUAGUUAUCCGGAUGGGCGCUACUUAAGCGCAGAAUCUGAUAGGGCAAUCGCACAAUAAUUUCAGUAAGAAGUCCAUCGUAGGACUCGAAAAUAUCCGGGUAAUU ...(((((((((((((.(((((.....))))).....((((...(((....)).).......))))...((.((((......)))))).....))))))))))))) ( -33.10, z-score = -3.22, R) >droSec1.super_3 3557618 106 + 7220098 CGUAGUUAUCCGGAUGGGCGCUACUUAAGCGCAGAAUCUGAUAGGGCAAUCGCACAAUAAUUUCAGUAAGAAGUCCAUCGUAGGACUCGAAAAUAUCCGGGUAAUU ...(((((((((((((.(((((.....))))).....((((...(((....)).).......))))...((.((((......)))))).....))))))))))))) ( -33.10, z-score = -3.22, R) >droYak2.chr2L 10690719 106 + 22324452 CGUAGUUAUCCGGAUGGGCGCUACUUAAGCGAAGGAUCUGAUACGGCAAUCGCACAAUAAUUUCAGUAAGAAGUCCAUCGUAAGAUUCGAAGAUAUCCGGGUAAUU ...(((((((((((((..((((.....))))..((((((..((((((....))......(((((.....)))))....)))))))))).....))))))))))))) ( -31.70, z-score = -2.63, R) >droEre2.scaffold_4929 16966259 106 + 26641161 CGUAAUUAUCCGGAUGGGCGCUACUUAAGCGAAGAAUCUGAUAAGGCAAUCGCACAAUAAUUUCAGUAAGAAGUCCAUCGUAGGACUCGAAAAUAUCCGGGUAGUU ...(((((((((((((..((((.....))))......((((....((....)).........))))...((.((((......)))))).....))))))))))))) ( -29.02, z-score = -2.25, R) >droAna3.scaffold_12916 11783666 106 - 16180835 CAUAGUUGUCCGGAUGGGCGCUGCUCAGGCGCAGGAUCUGGUACGGCAGUCGCACAAUAAUCUCUGUGAGGAGGGCGUCAUAGGACUCAAAAAUGUCCGGGUAGUU .......((((....))))(((((((.(((((..((.(((......))))).........(((((....))))))))))...((((........))))))))))). ( -35.10, z-score = -1.06, R) >dp4.chr4_group4 5572286 106 - 6586962 CGUAGUUGUCCGGAUGGGCGCUGCUUAGCCGCAGAAUCUGGUAGGGCAGCCGCACAAUGAUCUCCGUGAGCAGUCCGUCGUACGACUCAAAGAUAUCUGGAUAGUU .....((((((((((((((((((((..((((.......))))..)))))).((...(((.....)))..)).))))(((....)))........)))))))))).. ( -37.50, z-score = -1.23, R) >droPer1.super_46 42841 106 + 590583 CGUAGUUGUCCGGAUGGGCGCUGCUUAGCCGCAGAAUCUGGUAGGGCAGCCGCACAAUGAUCUCCGUGAGCAGUCCGUCGUACGACUCAAAGAUAUCUGGAUAGUU .....((((((((((((((((((((..((((.......))))..)))))).((...(((.....)))..)).))))(((....)))........)))))))))).. ( -37.50, z-score = -1.23, R) >droWil1.scaffold_180708 6303753 106 + 12563649 CAUAGUUAUCCGGAUGGGCACUACAAAGCCGCAAUAUCUGAUACGGCAGACGGACAAUAAUUUCCGUGAGUAGAGCAUCAUACGAUUCAAAUAUAUCAGGAUAGUU .....(((((((.((((((.((((...((((............))))..(((((........)))))..)))).)).)))).)(((........))).)))))).. ( -27.70, z-score = -2.64, R) >droVir3.scaffold_12963 16258868 106 + 20206255 CAUAGUUGUCCGGAUGGGCGCUGCUCAGGCGCAGAAUCUGAUACGGCAGACGCACAAUAAUCUCUGUGAGCAGCGCAUCAUACGACUCGAAUAUGUCCGGAUAGUU .....((((((((((..(((((((((..(((.....((((......)))))))(((........)))))))))))).((....)).........)))))))))).. ( -38.20, z-score = -2.04, R) >droMoj3.scaffold_6500 21199929 106 + 32352404 CAUAAUUAUCGGGAUGGGCGCUGCUCAGACGCAGUAUUUGGUAGGGCAGACGCACAAUAAUUUCGGUGAGCAGCGCAUCAUACGACUCAAAUAUGUCAGGAUAGUU ...((((((((..((((((((((((((..((.(((..(((.....((....)).)))..))).)).)))))))))).)))).)(((........)))..))))))) ( -34.20, z-score = -3.43, R) >droGri2.scaffold_15252 13684807 106 + 17193109 CAUAGUUGUCCGGGUGAGCGCUGCUCAGCCGCAAAAUCUGAUACGGUAGACGCACAAUGAUCUCGGUGAGCAGGCCGUCAUACGAUUCAAAAAUAUCCGGAUAGUU .....(((((((((((.((.(((((((.(((....(((.....((.....))......)))..))))))))))))((.....)).........))))))))))).. ( -33.50, z-score = -1.59, R) >anoGam1.chr2R 643756 97 + 62725911 --------GAUGGAUGUCCCGAAGAUAGUUUCAAAAUUUGGUAAGGUAGCCUAACCGCCAUCUCCGUUAGCACCUGUGAGUACAUCUCGAAUACAUC-GUUUGAUC --------....(((((..(((.(((.((((((..........((((.((.((((.(......).)))))))))).)))).))))))))...)))))-........ ( -18.10, z-score = 0.39, R) >triCas2.chrUn_77 25843 102 - 283703 --UAUCGGUUCGG-CAAUUUCCGAAUAUUUGU-GUACUUGAUACGGAAGUCGCAACGCCAUUUCGGUUAAAAUCUCCGGAUAAGUUGAAAAAAUAUCCGGACCGUU --...((((((((-..(((((((((.....((-((...((..((....))..))))))...)))))...))))..))(((((..((....)).))))))))))).. ( -27.10, z-score = -1.31, R) >consensus CGUAGUUAUCCGGAUGGGCGCUACUUAGGCGCAGAAUCUGAUACGGCAGUCGCACAAUAAUUUCAGUGAGCAGUCCAUCGUACGACUCAAAAAUAUCCGGAUAGUU .....(((((((((((((.....))).............(((......)))...........................................)))))))))).. (-11.66 = -10.16 + -1.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:59 2011