
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,678,472 – 15,678,574 |
| Length | 102 |
| Max. P | 0.847063 |

| Location | 15,678,472 – 15,678,574 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Shannon entropy | 0.42014 |
| G+C content | 0.34465 |
| Mean single sequence MFE | -18.33 |
| Consensus MFE | -12.88 |
| Energy contribution | -12.38 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15678472 102 - 27905053 CAGAGAGCGGCAAACAAAAAGAGGUAUUCAUUA--AAUGACUUAAAAUAAAUUGGGUUAAACUAAAUAUUUAGU-----AAUCAGUGCCAAAGCGCAUCGCUGUCUAUG .(((.(((((............(((........--..((((((((......)))))))).(((((....)))))-----.))).((((....)))).))))).)))... ( -22.00, z-score = -2.17, R) >droWil1.scaffold_181108 573315 108 + 4707319 -AAAAAACAGCACAUUAAGCAAAAAAACAACGACUAAAGGUUUAAGGACAAUCAAAUUAGUGGUAUGAAUUAAUGGCACAGCUAAUGCCACAAGGCAUCGCUGUCAAUG -........((.((((((.........((((.(((((.((((.......))))...))))).)).))..))))))))(((((..(((((....))))).)))))..... ( -21.60, z-score = -1.91, R) >droEre2.scaffold_4770 11761797 99 + 17746568 ---AAUGCGGCAAACAAAACGAGGGACUAAUUA--AACGACUUAAAAUAAAUUAGGUUAAUCUAAGUAAUCUGU-----AAUCAGUGCCAAAGCGCAUCGCUGUCAAUG ---...(((((.........((...((..((((--...(((((((......)))))))........))))..))-----..)).((((....))))...)))))..... ( -16.80, z-score = -0.48, R) >droYak2.chr3R 4449109 101 - 28832112 -CAAGAGCGGCAAACAAAACGAGGUAUUCAUUA--AAUGACUUAAAAUAAAUUAAGAUAAUCUAAAUAAUCAGU-----AAUCAGUGCCAAAGCGCAUCGCUGUCAAUG -...((.((((.........((..((((.((((--...(((((((......)))))....))....)))).)))-----).)).((((....))))...)))))).... ( -14.70, z-score = -0.60, R) >droSec1.super_25 728884 101 + 827797 -CAAAAGCGGCAAACAGAACGAGGUAUUCAUUA--AAUGACUUAAAAUAAAUUAAGUUAAACUAAAUAAUCAGU-----AAUCAGUGCCAAAGCGCAUCGCUGUCUAUG -.....(((((.........((..((((.((((--..((((((((......)))))))).......)))).)))-----).)).((((....))))...)))))..... ( -17.00, z-score = -1.30, R) >droSim1.chr3R 21721047 101 + 27517382 -CAAAAGCGGCAAACAAAAUGAGGUAUUCAUUA--AAUGACUUAAAAUAAAUUAAGUUAAACUAAACAAUCAGU-----AAUCAGUGCCAAAGCGCAUCGCUGUCUAUG -.....(((((..((..((((((...)))))).--..((((((((......)))))))).............))-----.....((((....))))...)))))..... ( -17.90, z-score = -1.89, R) >consensus _CAAAAGCGGCAAACAAAACGAGGUAUUCAUUA__AAUGACUUAAAAUAAAUUAAGUUAAACUAAAUAAUCAGU_____AAUCAGUGCCAAAGCGCAUCGCUGUCAAUG ......(((((..........................((((((((......)))))))).........................((((......)))).)))))..... (-12.88 = -12.38 + -0.49)

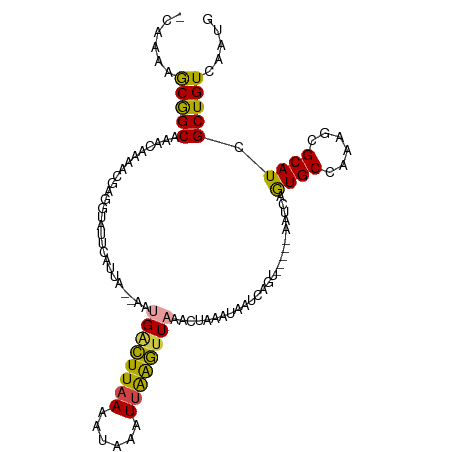
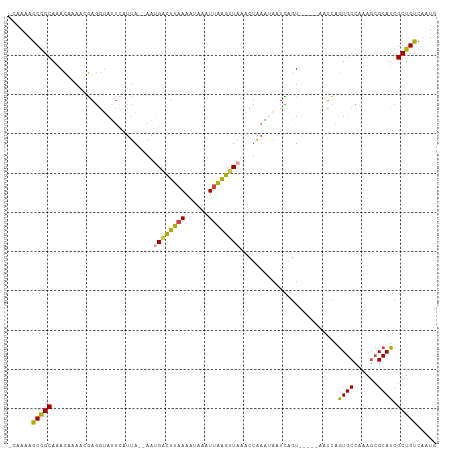
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:38 2011