| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,660,998 – 15,661,089 |
| Length | 91 |
| Max. P | 0.939720 |

| Location | 15,660,998 – 15,661,089 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 72.06 |
| Shannon entropy | 0.58236 |
| G+C content | 0.41178 |
| Mean single sequence MFE | -15.80 |
| Consensus MFE | -9.77 |
| Energy contribution | -9.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939720 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
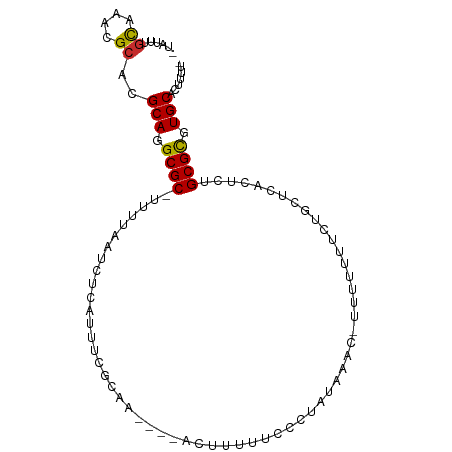
>dm3.chr3R 15660998 91 - 27905053 -UAUUUGCAAACGCACGCAGGCGC-UUUUAAUCUCAUUUCGCUA----ACUUUUUCGCAAUAAAC-UUUUUUUCUGCUCACUCUGCGCGUGCACUUUU- -...........((((((.((((.-..............)))).----........(((..(((.-...)))..))).........))))))......- ( -16.46, z-score = -0.83, R) >droSim1.chr3R 21703395 91 + 27517382 -UAUUUGCAAACGCACGCAGGCGC-UUUUAAUCUCAUUUCGCCA----ACUUUUUCGCUAUAAAC-UUUUUUUCUGCUCACUCCGCGCGUGCACUUUG- -...........((((((.((((.-..............)))).----.................-.........((.......))))))))......- ( -18.06, z-score = -1.61, R) >droSec1.super_25 711486 92 + 827797 -UAUUUGCAAACGCACGCAGGCGC-UUUUAAUCUCAUUUCGCCA----ACUUUUUCGCUAUAAACUUUUUUUUCUGCUCACUCCGCGCGUGCACUUUG- -...........((((((.((((.-..............)))).----...........................((.......))))))))......- ( -18.06, z-score = -1.60, R) >droYak2.chr3R 4430409 90 - 28832112 -UAUUUGCAAACGCACGCAGGCGC-UUUUAAUCUCAUUUCGCGA-----CUUUUUCCCUAUAAAC-UUUUUUCUUGCUCACUCUGCGCGUGCACUUUU- -...........((((((((((((-...............))).-----))).............-.........((.......))))))))......- ( -16.56, z-score = -1.36, R) >droEre2.scaffold_4770 11744439 89 + 17746568 -UAUUUGCAAACGCACGCAGGCGC-UUUUAAUCUCAUUUCACCA------CUUUUCCCUAUAAAC-UUUUUUUCUGCUCACGCUGCGCGUGCACUUUU- -...........((((((.((((.-...................------...............-..............))))..))))))......- ( -14.89, z-score = -1.16, R) >droAna3.scaffold_13340 2243675 89 - 23697760 -UAUUAUCAAACGUACGCAGGCGC-UUUUAAUCUCGAUUCGCAA----ACUAUUUCCCUAUAA----UUCAUUCUGCCCCGCUUGCGCGUGCACUUUUC -...........((((((..(((.-.((.......))..)))..----...............----........((.......))))))))....... ( -13.10, z-score = -0.27, R) >dp4.chr2 30185771 97 - 30794189 GUAUCUGCAAACGCACGCAGGCGCUUUUUAAUCUCAUUUGGCAAUACUUUUAUUUCACUUUUUC--UCCCCCUCUUUUUUUUCUGCGCGUGCAGUUUUU ....((((..((((..(((((.(((..............)))((((....))))..........--...............)))))))))))))..... ( -14.14, z-score = -0.59, R) >droPer1.super_6 5573618 98 - 6141320 GUAUCUGCAAACGCACGCAGGCGCUUUUUAAUCUCAUUUGGCAAUACUUUUAUUUCACUUUUUCU-CCCCCUCUUUUUUUUUCUGCGCGUGCAGUUUUU ....((((..((((..(((((.(((..............)))((((....))))...........-...............)))))))))))))..... ( -14.14, z-score = -0.53, R) >droWil1.scaffold_181108 555172 77 + 4707319 ------GUAUCCGCACGCACGCGCUGAUAAAUCUGAAAUAAUUU---UGUUCUUUUUCUAUG----UAUUUUU---------UUGCGUGUGCAAUUUUU ------..........((((((((.((.(((((.((((......---.......))))...)----.)))).)---------).))))))))....... ( -16.82, z-score = -2.23, R) >consensus _UAUUUGCAAACGCACGCAGGCGC_UUUUAAUCUCAUUUCGCAA____ACUUUUUCCCUAUAAAC_UUUUUUUCUGCUCACUCUGCGCGUGCACUUUU_ ......((....))..(((.((((............................................................)))).)))....... ( -9.77 = -9.68 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:35 2011