
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,631,768 – 15,631,829 |
| Length | 61 |
| Max. P | 0.896517 |

| Location | 15,631,768 – 15,631,829 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 85.27 |
| Shannon entropy | 0.30338 |
| G+C content | 0.43149 |
| Mean single sequence MFE | -12.24 |
| Consensus MFE | -8.64 |
| Energy contribution | -9.51 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
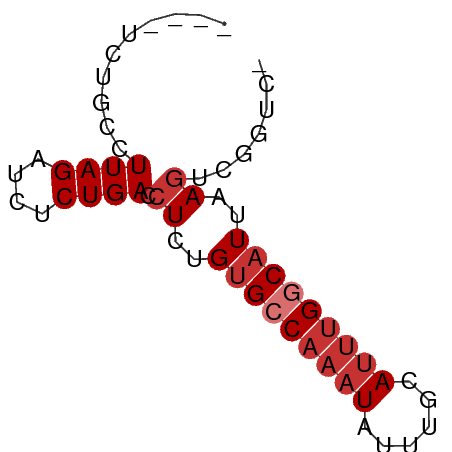
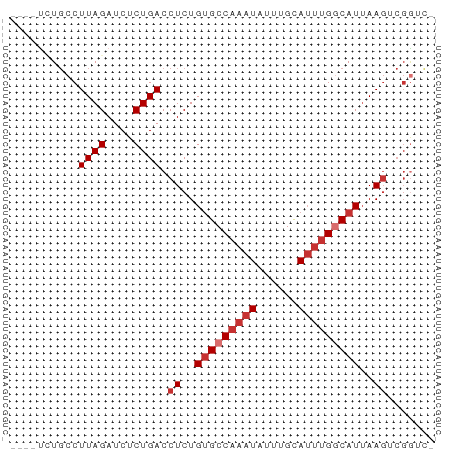
>dm3.chr3R 15631768 61 + 27905053 CUGCUCUGCCUUAGAUCUCUGACCUCUGUGCCAAAUAUUUGCAUUUGCCAUUAAGUCGGUC- ..................(((((....(((.(((((......))))).)))...)))))..- ( -9.10, z-score = -0.04, R) >droWil1.scaffold_181108 521119 54 - 4707319 CCACAUUCCCUUAGAUCUCUGACCUCUGUGCCAAAUAUUUGUAAACGGCAUUAA-------- ..........((((....)))).....(((((..............)))))...-------- ( -5.64, z-score = -0.33, R) >droPer1.super_6 5541788 59 + 6141320 ---UUAUGCCUUAGAUCUCUGACCUCUGUGCCAAAUAUUUGCAUUUGGCAUUAAGACGGAUC ---.......((((....))))((((((((((((((......)))))))))..))).))... ( -15.00, z-score = -2.42, R) >dp4.chr2 30153817 59 + 30794189 ---UUAUGCCUUAGAUCUCUGACCUCUGUGCCAAAUAUUUGCAUUUGGCAUUAAGACGGAUC ---.......((((....))))((((((((((((((......)))))))))..))).))... ( -15.00, z-score = -2.42, R) >droAna3.scaffold_13340 2213532 57 + 23697760 ----UCUGCCUUAGAUCUCUGACCUCUGUGCCAAAUAUUUGCAUUUGCCAUAAAGUCGAUC- ----.........((((...(((...((((.(((((......))))).))))..)))))))- ( -8.50, z-score = -0.96, R) >droEre2.scaffold_4770 11716170 57 - 17746568 ----CCUGCCUUAGAUCUCUGACCUCUGCGCCAAAUAUUUGCAUUUGGCAUUAAGUCGGUC- ----..............(((((......(((((((......))))))).....)))))..- ( -12.30, z-score = -1.09, R) >droYak2.chr3R 4394655 57 + 28832112 ----UCUGCCUUAGAUCUCUGACCUCUGUGCCAAAUAUUUGCAUUUGGCAUUAAGUCGGUC- ----..............(((((....(((((((((......)))))))))...)))))..- ( -16.20, z-score = -2.98, R) >droSec1.super_25 682773 57 - 827797 ----UCUGCCUUAGAUCUCUGACCUCUGUGCCAAAUAUUUGCAUUUGGCAUUAAGUCGGUC- ----..............(((((....(((((((((......)))))))))...)))))..- ( -16.20, z-score = -2.98, R) >consensus ____UCUGCCUUAGAUCUCUGACCUCUGUGCCAAAUAUUUGCAUUUGGCAUUAAGUCGGUC_ ..........((((....)))).((..(((((((((......)))))))))..))....... ( -8.64 = -9.51 + 0.88)

| Location | 15,631,768 – 15,631,829 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 85.27 |
| Shannon entropy | 0.30338 |
| G+C content | 0.43149 |
| Mean single sequence MFE | -14.76 |
| Consensus MFE | -9.71 |
| Energy contribution | -9.89 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
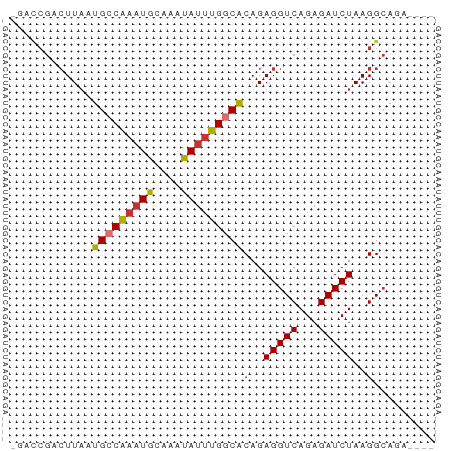
>dm3.chr3R 15631768 61 - 27905053 -GACCGACUUAAUGGCAAAUGCAAAUAUUUGGCACAGAGGUCAGAGAUCUAAGGCAGAGCAG -(.(((((((..((.((((((....)))))).))...)))))((....))..)))....... ( -12.40, z-score = -1.01, R) >droWil1.scaffold_181108 521119 54 + 4707319 --------UUAAUGCCGUUUACAAAUAUUUGGCACAGAGGUCAGAGAUCUAAGGGAAUGUGG --------....(((((..((....))..)))))(..(((((...)))))...)........ ( -8.40, z-score = -0.11, R) >droPer1.super_6 5541788 59 - 6141320 GAUCCGUCUUAAUGCCAAAUGCAAAUAUUUGGCACAGAGGUCAGAGAUCUAAGGCAUAA--- .....((((((.(((((((((....)))))))))....((((...))))))))))....--- ( -16.70, z-score = -2.79, R) >dp4.chr2 30153817 59 - 30794189 GAUCCGUCUUAAUGCCAAAUGCAAAUAUUUGGCACAGAGGUCAGAGAUCUAAGGCAUAA--- .....((((((.(((((((((....)))))))))....((((...))))))))))....--- ( -16.70, z-score = -2.79, R) >droAna3.scaffold_13340 2213532 57 - 23697760 -GAUCGACUUUAUGGCAAAUGCAAAUAUUUGGCACAGAGGUCAGAGAUCUAAGGCAGA---- -((((((((((.((.((((((....)))))).))..))))))...)))).........---- ( -13.60, z-score = -1.99, R) >droEre2.scaffold_4770 11716170 57 + 17746568 -GACCGACUUAAUGCCAAAUGCAAAUAUUUGGCGCAGAGGUCAGAGAUCUAAGGCAGG---- -(.(((((((..(((((((((....)))))))))...)))))((....))..)))...---- ( -16.30, z-score = -2.35, R) >droYak2.chr3R 4394655 57 - 28832112 -GACCGACUUAAUGCCAAAUGCAAAUAUUUGGCACAGAGGUCAGAGAUCUAAGGCAGA---- -(.(((((((..(((((((((....)))))))))...)))))((....))..)))...---- ( -17.00, z-score = -3.32, R) >droSec1.super_25 682773 57 + 827797 -GACCGACUUAAUGCCAAAUGCAAAUAUUUGGCACAGAGGUCAGAGAUCUAAGGCAGA---- -(.(((((((..(((((((((....)))))))))...)))))((....))..)))...---- ( -17.00, z-score = -3.32, R) >consensus _GACCGACUUAAUGCCAAAUGCAAAUAUUUGGCACAGAGGUCAGAGAUCUAAGGCAGA____ ............(((((((((....)))))))))(..(((((...)))))..)......... ( -9.71 = -9.89 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:33 2011