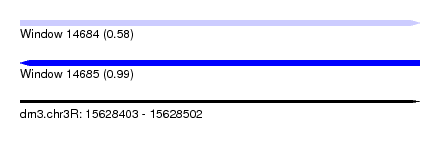
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,628,403 – 15,628,502 |
| Length | 99 |
| Max. P | 0.993972 |

| Location | 15,628,403 – 15,628,502 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Shannon entropy | 0.46066 |
| G+C content | 0.53645 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -25.38 |
| Energy contribution | -23.97 |
| Covariance contribution | -1.42 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
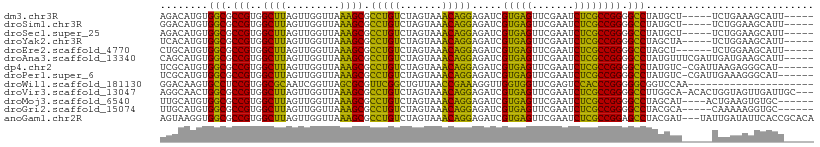
>dm3.chr3R 15628403 99 + 27905053 AGACAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUGCU-----UCUGAAAGCAUU----- (((((...(((((..(((((.....)))))..)))))))))).......(((...(((((((.......)))).)))..)))(((((-----(....)))))).----- ( -31.60, z-score = -0.62, R) >droSim1.chr3R 21657131 99 - 27517382 GGACAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUGCU-----UCUGGAAGCAUU----- (((((...(((((..(((((.....)))))..)))))))))).......(((...(((((((.......)))).)))..)))(((((-----(....)))))).----- ( -32.60, z-score = -0.46, R) >droSec1.super_25 679432 99 - 827797 AGACAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUGCU-----UCUGGAAGCAUU----- (((((...(((((..(((((.....)))))..)))))))))).......(((...(((((((.......)))).)))..)))(((((-----(....)))))).----- ( -32.60, z-score = -0.61, R) >droYak2.chr3R 4391263 99 + 28832112 UCACAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAGCUA-----UCUGGAAGCAUU----- ((.((.((((((((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))...))))-----).))))......----- ( -30.60, z-score = -0.10, R) >droEre2.scaffold_4770 11712915 98 - 17746568 CUGCAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAGCU------UCUGGAAGCAUU----- .(((....(((((..(((((.....)))))..)))))..(((((.....(((...(((((((.......)))).)))..)))....------.))))).)))..----- ( -31.60, z-score = -0.07, R) >droAna3.scaffold_13340 2210190 104 + 23697760 CAGCAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUGUUUCGAUUGAUGAAGCAUU----- ......((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))))((((((.....))))))...----- ( -31.40, z-score = -0.31, R) >dp4.chr2 30150382 102 + 30794189 UCGCAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUGUC-CGAUUAAGAGGGCAU------ ......((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))))(((-(........))))..------ ( -33.30, z-score = -0.34, R) >droPer1.super_6 5538374 102 + 6141320 UCGCAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUGUC-CGAUUGAAAGGGCAU------ ......((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))))(((-(........))))..------ ( -33.30, z-score = -0.37, R) >droWil1.scaffold_181130 2600403 88 - 16660200 GGACAAGUGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGUCCAA--------------------- ((((....(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).))))..--------------------- ( -42.60, z-score = -2.96, R) >droVir3.scaffold_13047 17154231 105 + 19223366 AGGCAACUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUUGGCA-ACACUGGUAGUUGAUUGC--- (((((...(((((..(((((.....)))))..)))))))))).((((.((((((((((......)).)))))(((((.((....)).-...)))))..))).))))--- ( -36.10, z-score = -0.68, R) >droMoj3.scaffold_6540 8815267 99 - 34148556 UUGCAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAGCAU----ACUGAAGUGUGC------ .......(((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).))))))((((----((....))))))------ ( -32.80, z-score = -0.67, R) >droGri2.scaffold_15074 2203512 98 - 7742996 UUGCAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUACGCA-----CAAAAAGGUGC------ ......((((.(((.((((((((..(((......)))...)))).......(((((((......)).))))))))).)))))))(((-----(......))))------ ( -33.80, z-score = -1.07, R) >anoGam1.chr2R 62199796 106 - 62725911 AGUAAGGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGAGCCUACGAU---UAUUGAUAUUCACCGCACA .((..((((((((..(((((.....)))))..)))))...........(((..(((((((.((((...........)))).))))))---).)))......)))..)). ( -30.40, z-score = -0.14, R) >consensus UGGCAUGUGGCGCCGUGGCUUAGUUGGUUAAAGCGCCUGUCUAGUAAACAGGAGAUCGUGAGUUCGAAUCUCGCCGGGGCCUAUGCU_____UCUGAAAGCAUU_____ ........(((.(((..((((.........)))).(((((.......))))).....(((((.......)))))))).)))............................ (-25.38 = -23.97 + -1.42)
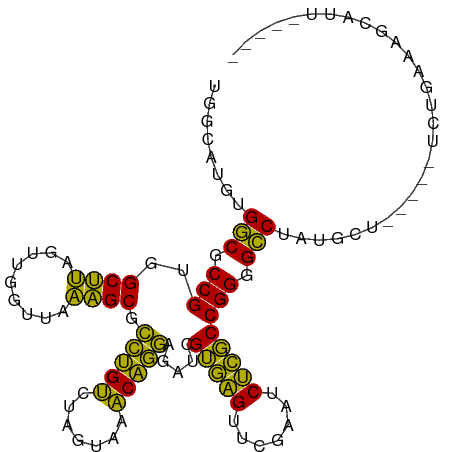
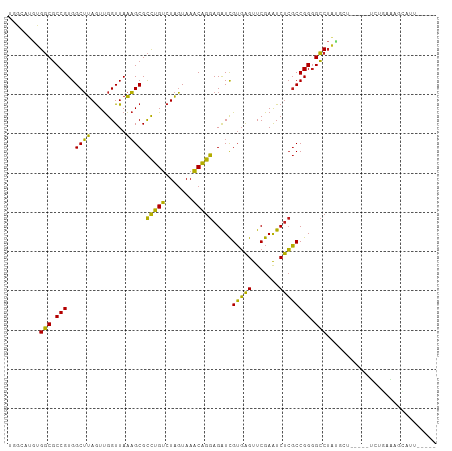
| Location | 15,628,403 – 15,628,502 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Shannon entropy | 0.46066 |
| G+C content | 0.53645 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -24.97 |
| Energy contribution | -24.36 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.993972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15628403 99 - 27905053 -----AAUGCUUUCAGA-----AGCAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGUCU -----.((((((....)-----))))).(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -30.20, z-score = -2.61, R) >droSim1.chr3R 21657131 99 + 27517382 -----AAUGCUUCCAGA-----AGCAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGUCC -----.((((((....)-----))))).(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -30.90, z-score = -2.92, R) >droSec1.super_25 679432 99 + 827797 -----AAUGCUUCCAGA-----AGCAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGUCU -----.((((((....)-----))))).(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -30.90, z-score = -2.82, R) >droYak2.chr3R 4391263 99 - 28832112 -----AAUGCUUCCAGA-----UAGCUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGUGA -----...(((......-----.)))..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -26.80, z-score = -1.17, R) >droEre2.scaffold_4770 11712915 98 + 17746568 -----AAUGCUUCCAGA------AGCUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGCAG -----...((((....)------)))..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -28.40, z-score = -1.94, R) >droAna3.scaffold_13340 2210190 104 - 23697760 -----AAUGCUUCAUCAAUCGAAACAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGCUG -----.(((.(((.......))).))).(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -26.70, z-score = -1.95, R) >dp4.chr2 30150382 102 - 30794189 ------AUGCCCUCUUAAUCG-GACAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGCGA ------(((.((........)-).))).(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -27.30, z-score = -1.22, R) >droPer1.super_6 5538374 102 - 6141320 ------AUGCCCUUUCAAUCG-GACAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGCGA ------(((.((........)-).))).(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -27.30, z-score = -1.28, R) >droWil1.scaffold_181130 2600403 88 + 16660200 ---------------------UUGGACCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCACUUGUCC ---------------------..((((.(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))....)))) ( -33.90, z-score = -2.47, R) >droVir3.scaffold_13047 17154231 105 - 19223366 ---GCAAUCAACUACCAGUGU-UGCCAAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAGUUGCCU ---(((((((((.......))-))....(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).))))).. ( -30.90, z-score = -1.99, R) >droMoj3.scaffold_6540 8815267 99 + 34148556 ------GCACACUUCAGU----AUGCUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGCAA ------..........((----(((...(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).))))).. ( -29.20, z-score = -2.40, R) >droGri2.scaffold_15074 2203512 98 + 7742996 ------GCACCUUUUUG-----UGCGUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGCAA ------((((......)-----)))((.(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))))...... ( -30.60, z-score = -2.34, R) >anoGam1.chr2R 62199796 106 + 62725911 UGUGCGGUGAAUAUCAAUA---AUCGUAGGCUCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACCUUACU ..((((((...........---))))))(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))........ ( -29.80, z-score = -1.60, R) >consensus _____AAUGCUUUCAGA_____AGCAUAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUGCCA ............................(((.(((((((((.(((......)))))))(((((.....)))))))((((.........))))..))).)))........ (-24.97 = -24.36 + -0.61)

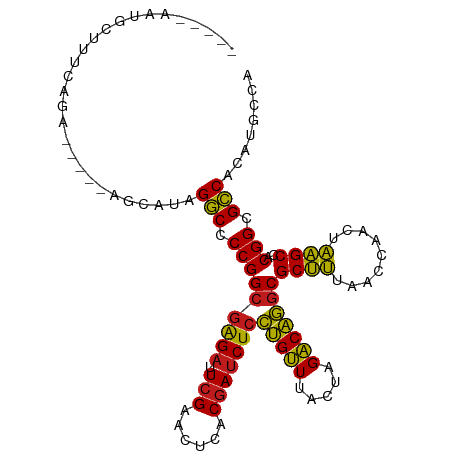
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:31 2011