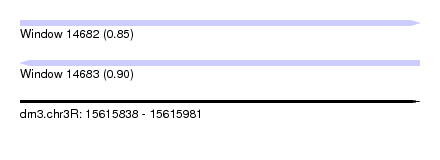
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,615,838 – 15,615,981 |
| Length | 143 |
| Max. P | 0.898348 |

| Location | 15,615,838 – 15,615,981 |
|---|---|
| Length | 143 |
| Sequences | 13 |
| Columns | 171 |
| Reading direction | forward |
| Mean pairwise identity | 68.87 |
| Shannon entropy | 0.63973 |
| G+C content | 0.44176 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.44 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.24 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
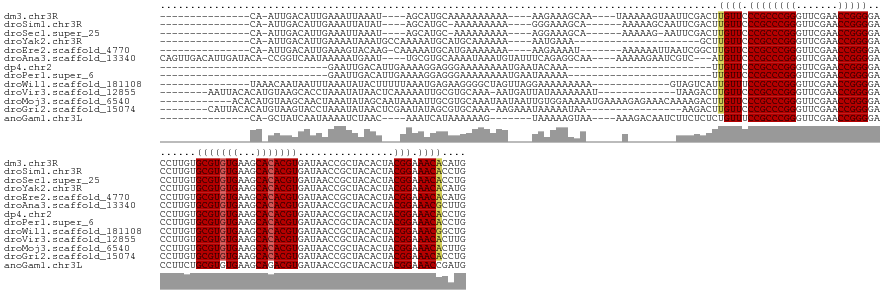
>dm3.chr3R 15615838 143 + 27905053 CAUGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGUCGAAUUACUUUUUA----UUGCUUUCUU----UUUUUUUUUUGCAUGCU----AUUUAAUUUCAAUGUCAAU-UG--------------- ((((((((((((..((((...(.......)(((((...)))))))))......(((((.......))))))))))))((((......))))....----..........----..........)))))..----...................-..--------------- ( -29.30, z-score = 0.47, R) >droSim1.chr3R 21644587 140 - 27517382 CAGGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGUCGAAUUGCUUUUU------UGCUUUCCC----UUUUUUUUU-GCAUGCU----AUAUAAUUUCAAUGUCAAU-UG--------------- ..((..((...((.((((((((........(((((...)))))(((.((((..(((((.......))))).(((((..(((.(((......)))------.))))))))----...)))).)-)).))))----)))).))...))..))...-..--------------- ( -35.60, z-score = -0.51, R) >droSec1.super_25 666954 139 - 827797 CAGGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGUCGAAUU-CUUUUU------UGCUUUCCU----UUUUUUUUU-GCAUGCU----AUUUAAUUUCAAUGUCAAU-UG--------------- ..((..((........((((((........(((((...)))))(((.((((..(((((.......))))).(((((..((.((((..-....))------)))))))))----...)))).)-)).))))----))........))..))...-..--------------- ( -30.59, z-score = 0.40, R) >droYak2.chr3R 4377797 130 + 28832112 CAUGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGC---------------------UUUCAUU----UUUUUUGCAUGCAUUUUUGGCAUUUAUUUUCAAUGUCAAU-UG--------------- ....((((((((..((((...(.......)(((((...)))))))))......(((((.......))))))))))))).((---------------------...((..----.....))...))....((((((((.......)))))))).-..--------------- ( -33.40, z-score = -0.63, R) >droEre2.scaffold_4770 11700675 143 - 17746568 CAUGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGCCGAUUAAUUUUUU-------AUUUUCUU----UUUUUUUCAUGCAUUUUUG-CUUGUACUUUCAAUGUCAAU-UG--------------- .(((((...((((......))))....)))))((((.......))))(((((.((((.((((....)))))))).((((((.((..........-------........----...............)).)-))))))))))..........-..--------------- ( -29.51, z-score = 0.45, R) >droAna3.scaffold_13340 2197882 159 + 23697760 CAAGCGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAU---GACGAUUCUUUUU----UUGCCUCUGAAAUACAUUUAUUUUGCACGCA----AUUCAUUUUUAUUGACCGG-UGUAUCAAUGUCAACUG .....(((((((..((((...(.......)(((((...)))))))))......(((((.......)))))))))))).(---((((.........----..((....((((((....))))))))((((.----..(((.......)))...)-))).....))))).... ( -36.40, z-score = 0.94, R) >dp4.chr2 30138967 119 + 30794189 CAGGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAA------------------------UUUGUAUUCAUUUUUUUUCCCUCCUUUUCAAUGUCAAUUC---------------------------- ...(((...((((......))))....)))(((((.....)))))..((((..(((((.......))))).(((((.((------------------------..((....))...)).))))).))))..............---------------------------- ( -30.40, z-score = -1.12, R) >droPer1.super_6 5527000 119 + 6141320 CAGGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAA------------------------UUUUUAUUCAUUUUUUUUCCCUCCUUUUCAAUGUCAAUUC---------------------------- ...(((...((((......))))....)))(((((.....)))))..((((..(((((.......))))).(((((.((------------------------.............)).))))).))))..............---------------------------- ( -29.02, z-score = -1.08, R) >droWil1.scaffold_181108 501161 143 - 4707319 CAGCCGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGAAACAAUGACUAC-------------UUUUUUUUUCCUAACUAGCCCCUUCUCAUUUAAAAAGUAUAUUUAAAUUAUUGUUUA--------------- .....(((((((..((((...(.......)(((((...)))))))))......(((((.......))))))))))))......(((-------------(((((..........................))))))))..................--------------- ( -29.07, z-score = 0.00, R) >droVir3.scaffold_12855 7296788 149 + 10161210 CAAGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGUCUUA-------------AUUUUUUUAUAAUCAUU-UUUGCACGCAAUUUUUGAGUUAUAUUUAGGUGCUUACAUGUGUAAUU-------- ...(((...((((......))))....)))(((((...)))))(((((.(((.((((.((((....)))))))).))((((((((-------------(......((((((((..-.(((....)))....))).))))).))))).)))).).)))))....-------- ( -36.80, z-score = -0.76, R) >droMoj3.scaffold_6540 8800539 159 - 34148556 CAAGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGUCUUUUGUUUCUCUUUUCAUUUUUCCACAAUUAUUAUUUGCACGCAAUUUUAUUGCUAUAUUUAGUUGCUUACAUGUGU------------ ...(((...((((......))))....)))(((((...)))))(((((.((..(((((.......))))).(((((((((...)))))))))............)).................((((((..((......))..)))))).....)))))------------ ( -37.80, z-score = -1.02, R) >droGri2.scaffold_15074 2188631 146 - 7742996 CAGGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGUCUU----------------UUAUUUUUAUUUCUU-UUUGCACGCUAUAUUCGAGUUAUAUUUAGGUACUUACAUGUGUAAUG-------- .((((((..(((.(((((((((........(((((...)))))(((.((((..(((((.......))))).(....).......----------------............)))-).))).))))))))))).)..))))))......((((....))))..-------- ( -36.00, z-score = -0.44, R) >anoGam1.chr3L 30142199 140 - 41284009 CAUCGGUUUCCGUAGUGUAGCGGUUAUCACGUCUGCUUCACACGCAGAAGGUCCCCGGUUCGAACCCGGGCGGAAACAGAGAGAAGAUUGUCUUU----UUACUUUUUA-------CUUUUUUAUGAUUU----GUUAGAUUUUAUUGAUAGC-UG--------------- .(((((((((((..(((.(((((.........))))).)))............(((((.......))))))))))))(((((((......)))))----))........-------..............----............))))...-..--------------- ( -32.30, z-score = 0.12, R) >consensus CAGGUGUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGUCGAA________U______UGCUUUUUA____UUUUUUUUUUGCAAUUUU___AUUAAAUUUCAAUGUCAAU_UG_______________ .....((((((.........(((..(((...(((((.......))))).)))..)))(((((....))))))))))).............................................................................................. (-25.71 = -25.44 + -0.27)

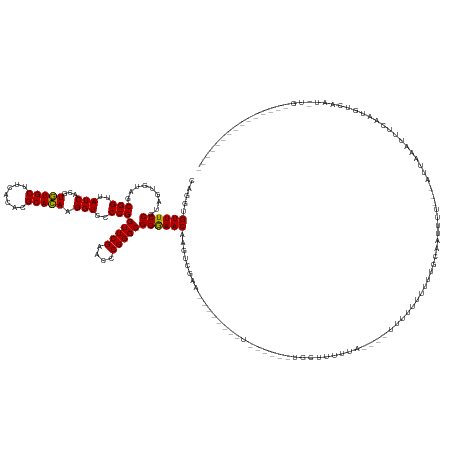
| Location | 15,615,838 – 15,615,981 |
|---|---|
| Length | 143 |
| Sequences | 13 |
| Columns | 171 |
| Reading direction | reverse |
| Mean pairwise identity | 68.87 |
| Shannon entropy | 0.63973 |
| G+C content | 0.44176 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -24.14 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.39 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15615838 143 - 27905053 ---------------CA-AUUGACAUUGAAAUUAAAU----AGCAUGCAAAAAAAAAA----AAGAAAGCAA----UAAAAAGUAAUUCGACUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACAUG ---------------..-..................(----(((..((((........----..........----....((((......))))(((((((..((....))...))))))).))))(((((((...))))))).......))))......(....)..... ( -27.60, z-score = 0.31, R) >droSim1.chr3R 21644587 140 + 27517382 ---------------CA-AUUGACAUUGAAAUUAUAU----AGCAUGC-AAAAAAAAA----GGGAAAGCA------AAAAAGCAAUUCGACUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACCUG ---------------..-..................(----(((..((-((.......----(((((....------...((....)).......))))).(((((.......)))))....))))(((((((...))))))).......))))......(....)..... ( -33.04, z-score = -0.35, R) >droSec1.super_25 666954 139 + 827797 ---------------CA-AUUGACAUUGAAAUUAAAU----AGCAUGC-AAAAAAAAA----AGGAAAGCA------AAAAAG-AAUUCGACUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACCUG ---------------..-..................(----(((....-........(----(((......------.....(-(((.......))))...(((((.......)))))..))))..(((((((...))))))).......))))......(....)..... ( -29.20, z-score = 0.12, R) >droYak2.chr3R 4377797 130 - 28832112 ---------------CA-AUUGACAUUGAAAAUAAAUGCCAAAAAUGCAUGCAAAAAA----AAUGAAA---------------------GCUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACAUG ---------------..-.....((((........((((.......))))........----))))...---------------------...((((.((((((((.......)))))....(((((((((((...))))))).))))...........))).)))).... ( -29.99, z-score = -0.34, R) >droEre2.scaffold_4770 11700675 143 + 17746568 ---------------CA-AUUGACAUUGAAAGUACAAG-CAAAAAUGCAUGAAAAAAA----AAGAAAAU-------AAAAAAUUAAUCGGCUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACAUG ---------------..-.............(((((((-((....)))..........----........-------..........((((.(((..(((....)))..))).)))).....))))))(((((...)))))(((....(((........)))...)))... ( -31.10, z-score = -0.44, R) >droAna3.scaffold_13340 2197882 159 - 23697760 CAGUUGACAUUGAUACA-CCGGUCAAUAAAAAUGAAU----UGCGUGCAAAAUAAAUGUAUUUCAGAGGCAA----AAAAAGAAUCGUC---AUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACGCUUG ..((((((.........-...)))))).....((((.----(((((.........))))).)))).((((..----..........(((---((((.....(((((.......)))))......((((.......)))))))))))..(((........)))....)))). ( -39.90, z-score = 0.10, R) >dp4.chr2 30138967 119 - 30794189 ----------------------------GAAUUGACAUUGAAAAGGAGGGAAAAAAAAUGAAUACAAA------------------------UUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACCUG ----------------------------..............((((.(((((....(((........)------------------------)).))))).(((((.......)))))..))))..(((((((...))))))).....(((........)))......... ( -32.20, z-score = -1.37, R) >droPer1.super_6 5527000 119 - 6141320 ----------------------------GAAUUGACAUUGAAAAGGAGGGAAAAAAAAUGAAUAAAAA------------------------UUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACCUG ----------------------------..............((((.(((((....(((........)------------------------)).))))).(((((.......)))))..))))..(((((((...))))))).....(((........)))......... ( -32.20, z-score = -1.56, R) >droWil1.scaffold_181108 501161 143 + 4707319 ---------------UAAACAAUAAUUUAAAUAUACUUUUUAAAUGAGAAGGGGCUAGUUAGGAAAAAAAAA-------------GUAGUCAUUGUUUCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACGGCUG ---------------....................((((((....)))))).(((((.((..........))-------------.))))).((((((((((((((.......)))))....(((((((((((...))))))).))))...........)))))))))... ( -36.60, z-score = -0.85, R) >droVir3.scaffold_12855 7296788 149 - 10161210 --------AAUUACACAUGUAAGCACCUAAAUAUAACUCAAAAAUUGCGUGCAAA-AAUGAUUAUAAAAAAAU-------------UAAGACUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACUUG --------.(((((........((((.....(((((.(((....(((....))).-..)))))))).......-------------........(((((((..((....))...)))))))...))))(((((...))))))))))..(((........)))......... ( -30.20, z-score = -0.00, R) >droMoj3.scaffold_6540 8800539 159 + 34148556 ------------ACACAUGUAAGCAACUAAAUAUAGCAAUAAAAUUGCGUGCAAAUAAUAAUUGUGGAAAAAUGAAAAGAGAAACAAAAGACUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACUUG ------------.....((((.(((..........(((((...))))).)))...........((((...........(((.(((((.....)))))....(((((.......)))))...)))..(((((((...))))))).....))))))))....(....)..... ( -33.60, z-score = -0.01, R) >droGri2.scaffold_15074 2188631 146 + 7742996 --------CAUUACACAUGUAAGUACCUAAAUAUAACUCGAAUAUAGCGUGCAAA-AAGAAAUAAAAAUAA----------------AAGACUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACCUG --------..((((....))))(((((((.((.........)).))).))))...-...............----------------......((((.((((((((.......)))))....(((((((((((...))))))).))))...........))).)))).... ( -30.20, z-score = -0.08, R) >anoGam1.chr3L 30142199 140 + 41284009 ---------------CA-GCUAUCAAUAAAAUCUAAC----AAAUCAUAAAAAAG-------UAAAAAGUAA----AAAGACAAUCUUCUCUCUGUUUCCGCCCGGGUUCGAACCGGGGACCUUCUGCGUGUGAAGCAGACGUGAUAACCGCUACACUACGGAAACCGAUG ---------------..-...................----..............-------..........----...............((.((((((((((((.......))))).....(((((.......))))).(((.....))).......))))))).)).. ( -28.30, z-score = -0.57, R) >consensus _______________CA_AUUGACAUUGAAAUUAAAU___AAAAAUGCAAAAAAAAAA____UAAAAAAAA______A________UUCGACUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAACACCUG .............................................................................................((((.((((((((.......)))))........(((((((...)))))))................))).)))).... (-24.14 = -24.18 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:30 2011