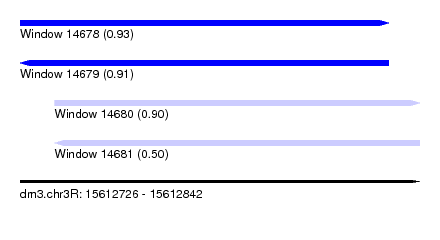
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,612,726 – 15,612,842 |
| Length | 116 |
| Max. P | 0.929053 |

| Location | 15,612,726 – 15,612,833 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 88.22 |
| Shannon entropy | 0.20899 |
| G+C content | 0.46042 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -23.16 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.929053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15612726 107 + 27905053 UUCCGAUCGGUGAUCGGUGAUCGGAGAUCGCAGAUCUCAGUUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUU .((((((((.(....).))))))))((((((((((..(.(((....))).)...))))))))))..........((((((......))))))(((((.....))))) ( -34.60, z-score = -2.08, R) >droEre2.scaffold_4770 11696146 107 - 17746568 UUCCGAUCGGCGAUUGGAGAUUGGAGAUCACAGAUCUCAGAUCCAGAAUCGCAAAUCUGCGAUCUAAUCCGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUU ....((((((((..((((((((.((((((...)))))).)))).(((.(((((....))))))))..))))..))))))))...........(((((.....))))) ( -34.70, z-score = -2.68, R) >droYak2.chr3R 4374736 93 + 28832112 --------------UUCCGAUCGGAGAUCGCAGAUCUCAGAUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUU --------------...(((.((((((((((((((....(((.....)))....))))))))))).........((((((......))))))..))))))....... ( -29.00, z-score = -2.81, R) >droSec1.super_1 14195114 107 - 14215200 UUCCGAUCGGUGAUCUGAGAUCGCAGAGCUCAGAUCACAGUUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCCGAUUAAUUCAUUAGCAACUGUCGACCAGUU ....(((((((((((((((.((...)).))))))))...((..((((((((((....))))))....)))).)))))))))...........(((((.....))))) ( -33.50, z-score = -2.70, R) >droSim1.chr3R 21641473 107 - 27517382 UUCCGAUCGGUGAUCUGAGAUCGCAGAGCUCAGAUCACAGUUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUU ...((((.(((((((((((.((...)).)))))))))).((..((((((((((....))))))....)))).))((((((......))))))..).))))....... ( -34.80, z-score = -2.82, R) >consensus UUCCGAUCGGUGAUCUGAGAUCGGAGAUCGCAGAUCUCAGUUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUU ...........(((((((((((...))))))))))).......((((((((((....))))))....))))...((((((......))))))(((((.....))))) (-23.16 = -24.60 + 1.44)

| Location | 15,612,726 – 15,612,833 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Shannon entropy | 0.20899 |
| G+C content | 0.46042 |
| Mean single sequence MFE | -36.84 |
| Consensus MFE | -25.52 |
| Energy contribution | -29.44 |
| Covariance contribution | 3.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15612726 107 - 27905053 AACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAACUGAGAUCUGCGAUCUCCGAUCACCGAUCACCGAUCGGAA ...((((((....((((...(((.....)))....))))...(((((((((((((.((.((....)).))))))))))))))).))))))((((((...)))))).. ( -36.40, z-score = -2.48, R) >droEre2.scaffold_4770 11696146 107 + 17746568 AACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCGGAUUAGAUCGCAGAUUUGCGAUUCUGGAUCUGAGAUCUGUGAUCUCCAAUCUCCAAUCGCCGAUCGGAA ..(((((((.(....(((.((......)).))).((.((((((((((((((((((..((((...))))..)))))))))))))..))))).))...).))))))).. ( -34.70, z-score = -1.70, R) >droYak2.chr3R 4374736 93 - 28832112 AACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAUCUGAGAUCUGCGAUCUCCGAUCGGAA-------------- ..(((((((....((((...(((.....)))....))))...(((((((((((((..((((...))))..))))))))))))).)))))))..-------------- ( -33.60, z-score = -2.64, R) >droSec1.super_1 14195114 107 + 14215200 AACUGGUCGACAGUUGCUAAUGAAUUAAUCGGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAACUGUGAUCUGAGCUCUGCGAUCUCAGAUCACCGAUCGGAA ..(((((((((((((((..((......))..))...((((....(((((......))))))))).))))))((((((((.((...)).))))))))..))))))).. ( -40.10, z-score = -3.13, R) >droSim1.chr3R 21641473 107 + 27517382 AACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAACUGUGAUCUGAGCUCUGCGAUCUCAGAUCACCGAUCGGAA ..((((((((((((((((.((......)).)))...((((....(((((......))))))))).))))))((((((((.((...)).))))))))..))))))).. ( -39.40, z-score = -2.97, R) >consensus AACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAACUGAGAUCUGAGAUCUCCGAUCUCAGAUCACCGAUCGGAA ..(((((((.((((((((.((......)).)))...((((....(((((......))))))))).))))).(((((((((((...)))))))))))..))))))).. (-25.52 = -29.44 + 3.92)

| Location | 15,612,736 – 15,612,842 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Shannon entropy | 0.32393 |
| G+C content | 0.43806 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.899027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15612736 106 + 27905053 UGAUCGGUGAUCGGAGAUCGCAGAUCUCAGUUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUUUGUCAGUUA (((((((((..((((((((((((((..(.(((....))).)...))))))))))...))))..)))))))))..........(((((.(((.....))).))))). ( -32.80, z-score = -2.10, R) >droEre2.scaffold_4770 11696156 106 - 17746568 CGAUUGGAGAUUGGAGAUCACAGAUCUCAGAUCCAGAAUCGCAAAUCUGCGAUCUAAUCCGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUUUGUCAGUUA .(((((((((((.((((((...)))))).)))).....(((((....)))))))))))).....((((((......))))))(((((.(((.....))).))))). ( -33.40, z-score = -2.86, R) >droYak2.chr3R 4374744 94 + 28832112 ------------GGAGAUCGCAGAUCUCAGAUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUUUGUCAGUUA ------------..(((((((((((....(((.....)))....))))))))))).........((((((......))))))(((((.(((.....))).))))). ( -29.70, z-score = -2.98, R) >droAna3.scaffold_13340 2194980 80 + 23697760 --------------------------ACAGAUCUGCGAUCGCGAAUCUGCGAUCUAAUCUCCGUGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUUUGUCAGUUA --------------------------..((((.((.(((((((....)))))))))))))....((((((......))))))(((((.(((.....))).))))). ( -21.00, z-score = -1.59, R) >droSec1.super_1 14195124 106 - 14215200 UGAUCUGAGAUCGCAGAGCUCAGAUCACAGUUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCCGAUUAAUUCAUUAGCAACUGUCGACCAGUUUGUCAGUUA (((((((((.((...)).)))))))))..((..((((((((((....))))))....)))).))((..((......))..))(((((.(((.....))).))))). ( -29.90, z-score = -1.98, R) >droSim1.chr3R 21641483 106 - 27517382 UGAUCUGAGAUCGCAGAGCUCAGAUCACAGUUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUUUGUCAGUUA (((((((((.((...)).)))))))))..((..((((((((((....))))))....)))).))((((((......))))))(((((.(((.....))).))))). ( -33.40, z-score = -2.69, R) >consensus UGAUC_GAGAUCGGAGAUCGCAGAUCACAGAUCCAGAAUCGCAAAUCUGCGAUCUAAUCUGCACGCUGAUUAAUUCAUUAGCAACUGUCGACCAGUUUGUCAGUUA ..................................(((.(((((....)))))))).........((((((......))))))(((((.(((.....))).))))). (-17.58 = -17.50 + -0.08)

| Location | 15,612,736 – 15,612,842 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Shannon entropy | 0.32393 |
| G+C content | 0.43806 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -16.83 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 15612736 106 - 27905053 UAACUGACAAACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAACUGAGAUCUGCGAUCUCCGAUCACCGAUCA ...(((((.(((((.....)))))(((.((......)).))))).)))...(((((((((((((.((.((....)).)))))))))))))))..((((...)))). ( -31.40, z-score = -1.89, R) >droEre2.scaffold_4770 11696156 106 + 17746568 UAACUGACAAACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCGGAUUAGAUCGCAGAUUUGCGAUUCUGGAUCUGAGAUCUGUGAUCUCCAAUCUCCAAUCG .........(((((.....)))))(((.((......)).))).((.((((((((((((((((((..((((...))))..)))))))))))))..))))).)).... ( -29.50, z-score = -1.40, R) >droYak2.chr3R 4374744 94 - 28832112 UAACUGACAAACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAUCUGAGAUCUGCGAUCUCC------------ ...(((((.(((((.....)))))(((.((......)).))))).)))...(((((((((((((..((((...))))..)))))))))))))..------------ ( -31.00, z-score = -2.83, R) >droAna3.scaffold_13340 2194980 80 - 23697760 UAACUGACAAACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCACGGAGAUUAGAUCGCAGAUUCGCGAUCGCAGAUCUGU-------------------------- ...(((...(((((.....)))))(((.((......)).))).)))(((((.((((((......))))))...)))))..-------------------------- ( -22.30, z-score = -1.71, R) >droSec1.super_1 14195124 106 + 14215200 UAACUGACAAACUGGUCGACAGUUGCUAAUGAAUUAAUCGGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAACUGUGAUCUGAGCUCUGCGAUCUCAGAUCA ...((((..(((((.....)))))((..((......))..)).(((((((((((((((((.(((........))).)))))))))))..))))))...)))).... ( -33.50, z-score = -1.93, R) >droSim1.chr3R 21641483 106 + 27517382 UAACUGACAAACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAACUGUGAUCUGAGCUCUGCGAUCUCAGAUCA ...((((..(((((.....)))))(((.((......)).))).(((((((((((((((((.(((........))).)))))))))))..))))))...)))).... ( -32.80, z-score = -1.71, R) >consensus UAACUGACAAACUGGUCGACAGUUGCUAAUGAAUUAAUCAGCGUGCAGAUUAGAUCGCAGAUUUGCGAUUCUGGAACUGAGAUCUGAGAUCUCCGAUCUC_GAUCA ...(((((.(((((.....)))))(((.((......)).))))).)))....((((((......)))))).................................... (-16.83 = -16.92 + 0.08)
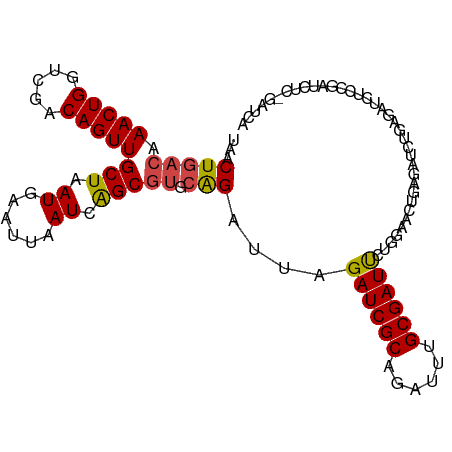
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:28 2011