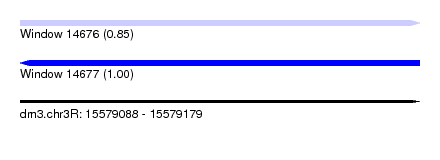
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,579,088 – 15,579,179 |
| Length | 91 |
| Max. P | 0.997137 |

| Location | 15,579,088 – 15,579,179 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.59 |
| Shannon entropy | 0.58433 |
| G+C content | 0.41847 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -7.85 |
| Energy contribution | -8.02 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
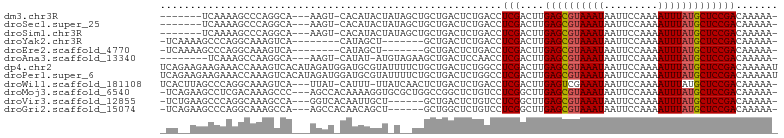
>dm3.chr3R 15579088 91 + 27905053 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGUCAGAGUCAGCAGCUAUAGUAUGUG-ACUU---UGCCUGGGCUUUUGA------- -.........((((.(((((.........))))).))))(((((.(((.((((((((.((((....)).))))-))))---))))).)))))....------- ( -28.70, z-score = -2.75, R) >droSec1.super_25 621558 91 - 827797 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGUCAGAGUCAGCAGCUAUAGUAUGUG-ACUU---UGCCUGGGCUUUUGA------- -.........((((.(((((.........))))).))))(((((.(((.((((((((.((((....)).))))-))))---))))).)))))....------- ( -28.70, z-score = -2.75, R) >droSim1.chr3R 21611768 91 - 27517382 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGUCAGAGUCAGCAGCUAUAGUAUGUG-ACUU---UGCCUGGGCUUUUGA------- -.........((((.(((((.........))))).))))(((((.(((.((((((((.((((....)).))))-))))---))))).)))))....------- ( -28.70, z-score = -2.75, R) >droYak2.chr3R 4342037 86 + 28832112 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGUCAGAGUCAGC-------AGCUAUG--------UGACUUUGCCUGGGCUUUUGA- -.........((((.(((((.........))))).))))(((((.(((.((((((((.(-------(....))--------))))))))))).)))))....- ( -28.80, z-score = -3.01, R) >droEre2.scaffold_4770 11666153 86 - 17746568 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGUCAGAGUCAGC-------AGCUAUG--------UGACUUUGCCUGGGCUUUUGA- -.........((((.(((((.........))))).))))(((((.(((.((((((((.(-------(....))--------))))))))))).)))))....- ( -28.80, z-score = -3.01, R) >droAna3.scaffold_13340 2160140 89 + 23697760 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGUUGGAGUCAGCUUCUACAU-AUAUG-ACUU---UGCCUUGGCUUUGA-------- -.........((((.(((((.........))))).))))(((((((((.(..(((((..........-...))-))).---.))))))))))...-------- ( -25.52, z-score = -2.72, R) >dp4.chr2 30100613 103 + 30794189 AUUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGCCAGAGUCAGCAGAAAAUACGCAUCCAUCUAUGUGACUUUGGUUUCUUCUUCUGA ..........((((.(((((.........))))).))))(((..(((((((((((((.((...................)))))))))))))))..))).... ( -26.11, z-score = -1.57, R) >droPer1.super_6 5479790 103 + 6141320 AUUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGCCAGAGUCAGCAGAAAAUACGCAUCCAUCUAUGUGACUUUGGUUUCUUCUUCUGA ..........((((.(((((.........))))).))))(((..(((((((((((((.((...................)))))))))))))))..))).... ( -26.11, z-score = -1.57, R) >droWil1.scaffold_181108 465865 97 - 4707319 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUCGACUCAAGUCGAGGUCAGAGUCAGAGUUGAUAA-AAAUG-AUAA---UGACUUUGCCUGGGCUAAGUGA -.....(((((((....((((....)))).)))))))...((((.(((.((((((((..(((.....-....)-))..---))))))))))).))))...... ( -25.60, z-score = -1.93, R) >droMoj3.scaffold_6540 8758164 98 - 34148556 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGCCGAGGACAGAGCCGGCCAGCGCACCUUUUGUGGCU---GGGCUUUGUCGAGGCUUCUGA- -.........((((.(((((.........))))).))))(((((...(((((((((...((((.(((.....))))))---))))))))))..)))))....- ( -36.30, z-score = -2.29, R) >droVir3.scaffold_12855 7240142 92 + 10161210 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGCCGAGGACAGAGUCAGC------AGCAAUUGUGACC---UGGCUUUGCCUGGGCUUCAGA- -.........((((.(((((.........))))).))))(((((.(((.(((((((((.------.((....))...)---))))))))))).)))))....- ( -33.80, z-score = -3.23, R) >droGri2.scaffold_15074 2142456 92 - 7742996 -UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGCCGAGGACAGAGCCAGC------AGCUGUUGUGGCU---UGGCUUUGCCUGGGCUUCUGA- -.........((((.(((((.........))))).))))(((((.(((.((((((((((------.((....)).)).---))))))))))).)))))....- ( -37.10, z-score = -3.23, R) >consensus _UUUUUGUCGGAGCAUAAAUUUUGGAAUUAUUUACGCUCAAGUCGAGGUCAGAGUCAGCAG__A_AACAUAUG_ACCU___UGACUUUGCCUUGGCUU__GA_ ..(((((...((((.(((((.........))))).))))....)))))....................................................... ( -7.85 = -8.02 + 0.17)

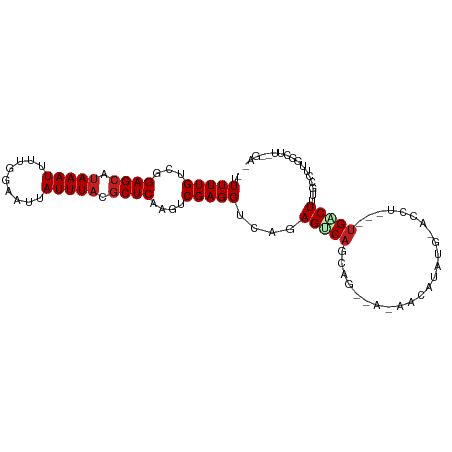
| Location | 15,579,088 – 15,579,179 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.59 |
| Shannon entropy | 0.58433 |
| G+C content | 0.41847 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -9.79 |
| Energy contribution | -9.88 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15579088 91 - 27905053 -------UCAAAAGCCCAGGCA---AAGU-CACAUACUAUAGCUGCUGACUCUGACCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- -------....(((.(.(((((---.(((-((((.........)).))))).)).))).).)))((((((((((.........)))))))))).........- ( -20.40, z-score = -3.09, R) >droSec1.super_25 621558 91 + 827797 -------UCAAAAGCCCAGGCA---AAGU-CACAUACUAUAGCUGCUGACUCUGACCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- -------....(((.(.(((((---.(((-((((.........)).))))).)).))).).)))((((((((((.........)))))))))).........- ( -20.40, z-score = -3.09, R) >droSim1.chr3R 21611768 91 + 27517382 -------UCAAAAGCCCAGGCA---AAGU-CACAUACUAUAGCUGCUGACUCUGACCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- -------....(((.(.(((((---.(((-((((.........)).))))).)).))).).)))((((((((((.........)))))))))).........- ( -20.40, z-score = -3.09, R) >droYak2.chr3R 4342037 86 - 28832112 -UCAAAAGCCCAGGCAAAGUCA--------CAUAGCU-------GCUGACUCUGACCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- -....(((.(.(((((.(((((--------((....)-------).))))).)).))).).)))((((((((((.........)))))))))).........- ( -21.20, z-score = -3.32, R) >droEre2.scaffold_4770 11666153 86 + 17746568 -UCAAAAGCCCAGGCAAAGUCA--------CAUAGCU-------GCUGACUCUGACCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- -....(((.(.(((((.(((((--------((....)-------).))))).)).))).).)))((((((((((.........)))))))))).........- ( -21.20, z-score = -3.32, R) >droAna3.scaffold_13340 2160140 89 - 23697760 --------UCAAAGCCAAGGCA---AAGU-CAUAU-AUGUAGAAGCUGACUCCAACCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- --------.....((....)).---..((-(....-......(((.(((........))).)))((((((((((.........)))))))))).))).....- ( -17.20, z-score = -2.32, R) >dp4.chr2 30100613 103 - 30794189 UCAGAAGAAGAAACCAAAGUCACAUAGAUGGAUGCGUAUUUUCUGCUGACUCUGGCCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAAU ....(((..((..(((.(((((((.(((((......)))))..)).))))).)))..))..)))((((((((((.........)))))))))).......... ( -23.00, z-score = -1.84, R) >droPer1.super_6 5479790 103 - 6141320 UCAGAAGAAGAAACCAAAGUCACAUAGAUGGAUGCGUAUUUUCUGCUGACUCUGGCCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAAU ....(((..((..(((.(((((((.(((((......)))))..)).))))).)))..))..)))((((((((((.........)))))))))).......... ( -23.00, z-score = -1.84, R) >droWil1.scaffold_181108 465865 97 + 4707319 UCACUUAGCCCAGGCAAAGUCA---UUAU-CAUUU-UUAUCAACUCUGACUCUGACCUCGACUUGAGUCGAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- ......(((...((((.(((((---....-.....-..........))))).)).))(((((....)))))..................)))..........- ( -14.01, z-score = -1.00, R) >droMoj3.scaffold_6540 8758164 98 + 34148556 -UCAGAAGCCUCGACAAAGCCC---AGCCACAAAAGGUGCGCUGGCCGGCUCUGUCCUCGGCUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- -....(((((..((((.(((((---((((((.....))).))))...)))).))))...)))))((((((((((.........)))))))))).........- ( -35.60, z-score = -4.27, R) >droVir3.scaffold_12855 7240142 92 - 10161210 -UCUGAAGCCCAGGCAAAGCCA---GGUCACAAUUGCU------GCUGACUCUGUCCUCGGCUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- -....(((((.(((......((---(((((((.....)------).)))).))).))).)))))((((((((((.........)))))))))).........- ( -25.50, z-score = -2.29, R) >droGri2.scaffold_15074 2142456 92 + 7742996 -UCAGAAGCCCAGGCAAAGCCA---AGCCACAACAGCU------GCUGGCUCUGUCCUCGGCUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA- -....(((((.(((((.(((((---(((.......)))------..))))).)).))).)))))((((((((((.........)))))))))).........- ( -32.10, z-score = -4.22, R) >consensus _UC__AAGCCAAAGCAAAGUCA___AGGU_CAUAUGCU_U__CUGCUGACUCUGACCUCGACUUGAGCGUAAAUAAUUCCAAAAUUUAUGCUCCGACAAAAA_ .........................................................(((....((((((((((.........)))))))))))))....... ( -9.79 = -9.88 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:25 2011