
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,560,274 – 15,560,368 |
| Length | 94 |
| Max. P | 0.603704 |

| Location | 15,560,274 – 15,560,368 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 89.97 |
| Shannon entropy | 0.19145 |
| G+C content | 0.34970 |
| Mean single sequence MFE | -15.91 |
| Consensus MFE | -14.77 |
| Energy contribution | -14.47 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
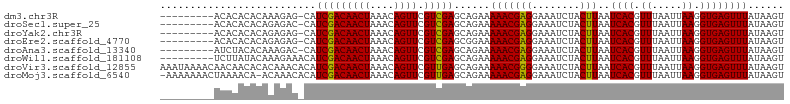
>dm3.chr3R 15560274 94 + 27905053 ---------ACACACACAAAGAG-CAUCGACAACUAAACAGUUCGUCGAGCAGAAAAACGAGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU ---------.............(-(.(((((((((....)))).)))))))....(((((((........)))..((((.((.....)).))))))))...... ( -18.10, z-score = -1.91, R) >droSec1.super_25 608098 94 - 827797 ---------ACACACACAGAGAC-CAUCGACAACUAAACAGUUCGUCGAGCAGAAAAACGAGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU ---------..((.((((((..(-(.(((((((((....)))).)))))...(.....)..))...))).((((((........))))))))).))........ ( -15.10, z-score = -0.57, R) >droYak2.chr3R 4327626 94 + 28832112 ---------ACACACACAGAGAG-CAUCGACAACUAAACAGUUCGUCGAGCAGAAAAACGAGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU ---------.............(-(.(((((((((....)))).)))))))....(((((((........)))..((((.((.....)).))))))))...... ( -18.10, z-score = -1.64, R) >droEre2.scaffold_4770 11653283 94 - 17746568 ---------ACACACACAGAGAG-CAUCGACAACUAAACAGUUCGUCGAGCGGAAAAACGAGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU ---------.............(-(.(((((((((....)))).)))))))....(((((((........)))..((((.((.....)).))))))))...... ( -18.10, z-score = -1.26, R) >droAna3.scaffold_13340 2145710 94 + 23697760 ---------AUCUACACAAAGAC-CAUCGACAACUAAACAGUUCGUCGAGCAGAAAAACGAGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU ---------..............-..(((((((((....)))).)))))......(((((((........)))..((((.((.....)).))))))))...... ( -15.10, z-score = -0.56, R) >droWil1.scaffold_181108 450496 95 - 4707319 ---------UCUUAUACAAAGAAACAUCGACAACUAAACAGUUCGUCGAGCAGAAAAACGAGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU ---------.(((((...........(((((((((....)))).)))))......(((((((........)))..((((.((.....)).))))))))))))). ( -17.00, z-score = -1.37, R) >droVir3.scaffold_12855 7219607 104 + 10161210 AAAUAAAACAACAACACACAAACACAUCGACAACUAAACAGUUCGUUGAGCAGAAAAACGGGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU ............(((.(((.......(((((((((....)))).))))).....................((((((........))))))))).)))....... ( -12.80, z-score = -0.26, R) >droMoj3.scaffold_6540 8736980 102 - 34148556 -AAAAAAACUAAAACA-ACAAACACAUCGACAACUAAACAGUUCGUUGAGCAGAAAAACGAGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU -...............-.........(((((((((....)))).)))))......(((((((........)))..((((.((.....)).))))))))...... ( -13.00, z-score = -0.32, R) >consensus _________ACACACACAAAGAC_CAUCGACAACUAAACAGUUCGUCGAGCAGAAAAACGAGGAAAUCUACUUAAUCACGUUUAAUUAAGGUGAGUUUAUAAGU ..........................(((((((((....)))).)))))......(((((((........)))..((((.((.....)).))))))))...... (-14.77 = -14.47 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:23 2011