
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,539,757 – 15,539,861 |
| Length | 104 |
| Max. P | 0.527670 |

| Location | 15,539,757 – 15,539,861 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.34 |
| Shannon entropy | 0.55197 |
| G+C content | 0.47824 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
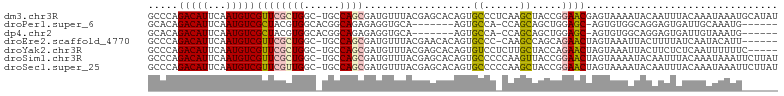
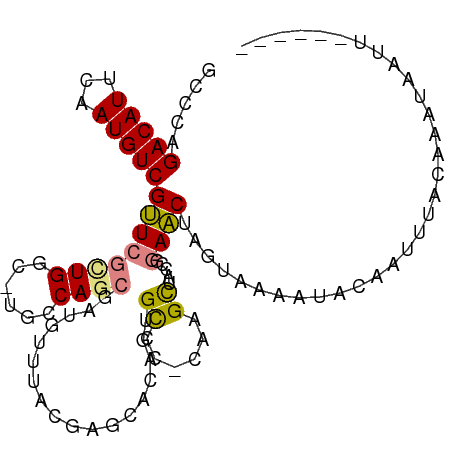
>dm3.chr3R 15539757 104 - 27905053 GCCCAGACAUUCAAUGUCGUUCGCUGGC-UGCCAGCGAUGUUUACGAGCACAGUGCCCUCAAGCUACCGGAACGAGUAAAAUACAAUUUACAAAUAAAUGCAUAU ((((.(((((...)))))..(((((((.-..))))))).((((..(.((.....)).)..))))....)).....(((((......)))))........)).... ( -21.30, z-score = -0.13, R) >droPer1.super_6 5432270 90 - 6141320 GCACAGACAUUCAAUGUCGCUACGUGGCACGGCAGAGAGGUGCA-------AGUGCCA-CCAGCAGCUGGAGC-AGUGUGGCAGGAGUGAUUGCAAAUG------ (((....(((((..((((((.(((((((((.(((......))).-------.))))))-).....((....))-.)))))))).)))))..))).....------ ( -31.70, z-score = -0.79, R) >dp4.chr2 30051084 90 - 30794189 GCACAGACAUUCAAUGUCGCUACGUGGCACGGCAGAGAGGUGCA-------AGUGCCA-CCAGCAGCUGGAGC-AGUGUGGCAGGAGUGAUUGUAAAUG------ ..((((.(((((..((((((.(((((((((.(((......))).-------.))))))-).....((....))-.)))))))).))))).)))).....------ ( -32.50, z-score = -1.41, R) >droEre2.scaffold_4770 11633033 97 + 17746568 GCCCAGACAUUCAAUGUCGUUCGCUGGC-UGCCAGCGAUGUUUACGAACACAGUGCCC-CAAGCCAGCAGAACUAGUAAAUUACUUUUAUCAAUACAUU------ .....(((((...)))))((((((((((-(....(((.(((........))).)))..-..))))))).))))..........................------ ( -25.50, z-score = -3.09, R) >droYak2.chr3R 4305749 99 - 28832112 GCCCAGACAUUCAAUGUCGUUCGCUGGC-UGCCAGCGAUGUUUACGAGCACAGUGUCCUCUUGCUACCAGAACUAGUAAAUUACUUCUCUCAAUUUUUUC----- .....((((((...(((((((((((((.-..))))))).....)))).)).))))))...((((((.......)))))).....................----- ( -20.60, z-score = -0.87, R) >droSim1.chr3R 21577993 104 + 27517382 GCCCAGACAUUCAAUGUCGUUCGCUGGC-UGCCAGCGAUGUUUACGAGCACAGUGCCCCCAAGUUACCGGAACUAGUAAAAUACAAUUUACAAAUAAAUUCUUAU ((.(.(((((...)))))..(((((((.-..))))))).......).))..(((.((...........)).))).(((((......))))).............. ( -19.40, z-score = -0.52, R) >droSec1.super_25 587789 104 + 827797 GCCCAGACAUUCAAUGUCGUUCGUUGGC-UGCCAGCGAUGUUUACGAGCACAGUGCCCCCAAGCUACCGGAACUAGUAAAAUACAAUUUACAAAUAAAUUCUUAU .....(((((...))))).((((.((((-(....(((.(((........))).))).....))))).))))....(((((......))))).............. ( -18.20, z-score = -0.10, R) >consensus GCCCAGACAUUCAAUGUCGUUCGCUGGC_UGCCAGCGAUGUUUACGAGCACAGUGCCC_CAAGCUACCGGAACUAGUAAAAUACAAUUUACAAAUAAUU______ .....(((((...)))))..(((((((....)))))))..(((((.......((((......)).))........)))))......................... (-12.32 = -12.35 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:22 2011