
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,512,595 – 15,512,693 |
| Length | 98 |
| Max. P | 0.517804 |

| Location | 15,512,595 – 15,512,693 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 67.59 |
| Shannon entropy | 0.62566 |
| G+C content | 0.55192 |
| Mean single sequence MFE | -30.01 |
| Consensus MFE | -11.71 |
| Energy contribution | -12.29 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15512595 98 + 27905053 GAAAUCAGCCAGCGAAUCAGCCAGUUUUUCAGUAACUGGCAAGCGUUCGAGCUGAGCGGG-CUAAGCCGGGCCCAGAGUGACAACAACAACAA------CGUGAG ....(((((...(((((..(((((((.......)))))))....))))).)))))(((((-((......)))))...((....))........------.))... ( -30.90, z-score = -1.64, R) >droSim1.chr3R 21559488 98 - 27517382 GAAGUCAGCCAGCGAGUCAGCCAGUUUUUCAGUAACUGGCAAGCGUUCGAGCUGAGCGGG-CUAAGCCGGGCCCAGAGUGACAAGAACAACAA------CGUGAG ...((((((((((..((..(((((((.......)))))))..))......)))).))(((-((......)))))....))))...........------(....) ( -34.00, z-score = -2.02, R) >droSec1.super_25 569340 98 - 827797 GAAGUCAGCCAGCGAGUCAGCCAGUUUUUCAGUAACUGGCAAGCGUUCGAGCUGAGCGGG-CUAAGCCGGGCCCAGAGUGACAAGAACAACAA------CGUGAG ...((((((((((..((..(((((((.......)))))))..))......)))).))(((-((......)))))....))))...........------(....) ( -34.00, z-score = -2.02, R) >droYak2.chr3R 4287177 104 + 28832112 GAAGUCAGCCAGCGAGUCAGCCAGUUUUUCAGUAACUGGCAAGCGUUCGAGCUGAACGGG-CUAAGCCGGGCCCAAAGUAACAAGAACAACAAGAACAACGUGAG ....((((((((((.....(((((((.......)))))))...)))).).)))))..(((-((......))))).........................(....) ( -29.90, z-score = -1.48, R) >droEre2.scaffold_4770 11615375 80 - 17746568 GAAGUCAGCCAGCGAGUGCGCCAGUUUUUCAGUAACUGGCAAGCGUUCGAGCUGAGCGGG-CUAAGGAACAAC-AACGUGGG----------------------- ..((((.(((((((((((((((((((.......)))))))..))))))..)))).)).))-))..........-........----------------------- ( -26.50, z-score = -0.95, R) >droAna3.scaffold_13340 2096946 95 + 23697760 GAAGCGAG---GGCAGCUUUUCAGCGACUCAGUAACUGGCUAGCGUUCAAGCUGAGCGGG-CUAAGCCGGGCCCCAAACAACGCAAAGGAGCA------AGUGAG ...(((.(---(((.(((....)))..........(((((((((((((.....))))..)-)).))))))))))).......((......)).------.))... ( -28.30, z-score = 0.91, R) >dp4.chr2 30025391 76 + 30794189 AAAACGCAAAGGCGAGAGCUCGAGGCUUUCAGUAGCUGAAAGUCGUUCGACUAGAGCGGGGCUAAGACCCAGUGGA----------------------------- ....(((...(((...(((((..(((((((((...)))))))))((((.....)))).)))))..).))..)))..----------------------------- ( -26.50, z-score = -1.47, R) >consensus GAAGUCAGCCAGCGAGUCAGCCAGUUUUUCAGUAACUGGCAAGCGUUCGAGCUGAGCGGG_CUAAGCCGGGCCCAAAGUGACAAGAACAACAA______CGUGAG ...................(((((((.......)))))))...(((((.....)))))............................................... (-11.71 = -12.29 + 0.57)

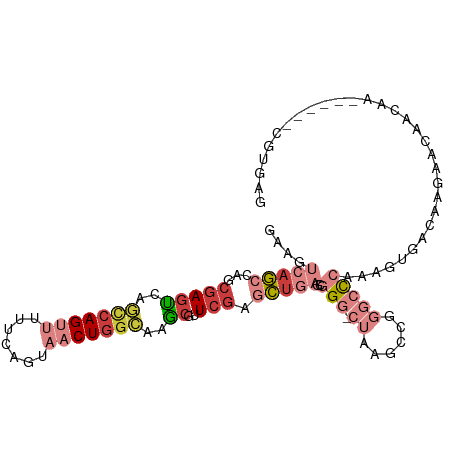
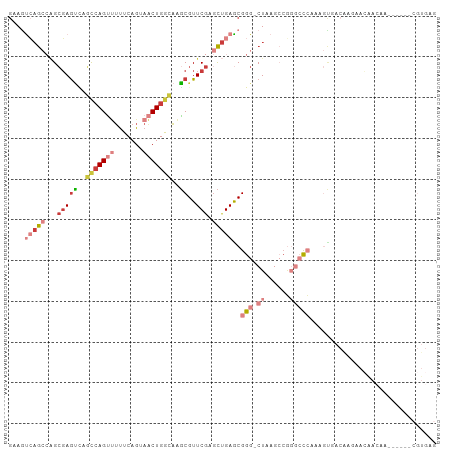
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:19 2011