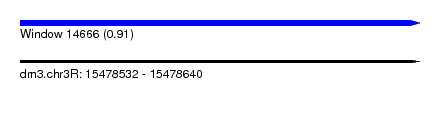
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,478,532 – 15,478,640 |
| Length | 108 |
| Max. P | 0.912235 |

| Location | 15,478,532 – 15,478,640 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 64.67 |
| Shannon entropy | 0.68207 |
| G+C content | 0.46155 |
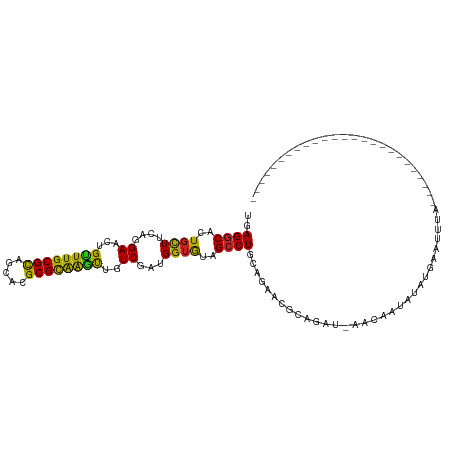
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -17.28 |
| Energy contribution | -16.52 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.43 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
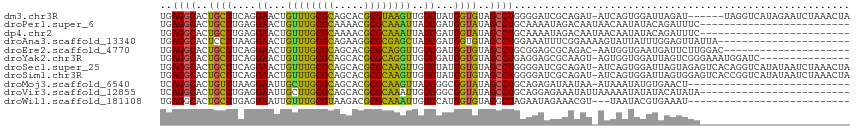
>dm3.chr3R 15478532 108 + 27905053 UGAGGCACUGCUUCAGGAACUGUUUGCGCAGCACGCGUAAGUUGUCUAUGGUGUAGCCUGGGGAUCGCAGAU-AUCAGUGGAUUAGAU------UAGGUCAUAGAAUCUAAACUA ((((((...))))))(((.(..(((((((.....)))))))..)((((((.....((((((.((((.((...-.....))))))...)------))))))))))).)))...... ( -26.90, z-score = 0.76, R) >droPer1.super_6 5365612 90 + 6141320 UGAGGCACUGCUUGAGGAACUGUUUGCGCAAAACGCGCAAAUUAUCGAUGGUAUAGCCUGCAAAAUAGACAAUAACAAUAUACAGAUUUC------------------------- ((((((..((((...((....((((((((.....))))))))..))...))))..)))).))............................------------------------- ( -20.40, z-score = -1.37, R) >dp4.chr2 29985346 90 + 30794189 UGAGGCACUGCUUGAGGAACUGUUUGCGCAAAACGCGCAAAUUAUCGAUGGUAUAGCCUGCAAAAUAGACAAUAACAAUAUACAGAUUUC------------------------- ((((((..((((...((....((((((((.....))))))))..))...))))..)))).))............................------------------------- ( -20.40, z-score = -1.37, R) >droAna3.scaffold_13340 2062457 93 + 23697760 UGAGGCACUCCUUAAGGAACUGUUUGCGCAGAAGGCGCGAGCUAUCGAUGGUGUAGCCUGGAAAUUUCGGAAAAGUAUUAUUUGGAGUUAUUA---------------------- (.((((((.((....((....((((((((.....))))))))..))...)).)).)))).).(((((((((.........)))))))))....---------------------- ( -25.30, z-score = -0.94, R) >droEre2.scaffold_4770 11581273 93 - 17746568 UGAGGCACUGCUUCAGGAACUGUUUGCGCAGCACGCGCAGGUUGUCGAUGGUGUAGCCUGCGGAGCGCAGAC-AAUGGUGAAUGAUUCUUGGAC--------------------- ((((((...)))))).....(((((((((....(.(((((((((.((....)))))))))))).))))))))-)....................--------------------- ( -31.50, z-score = -0.25, R) >droYak2.chr3R 4249856 99 + 28832112 UGAGGCACUGCUUCAGGAACUGUUUGCGCAGCACGCGCAGGUUGUCGAUGGUGUAGCCUGAGGAGCGCAAGU-AGUGGUGGAUUAGUCGGGAAAUGGAUC--------------- (((..(((..(((((((.(((((((((((.....))))).......))))))....)))))...((....))-))..)))..)))...............--------------- ( -27.61, z-score = 0.86, R) >droSec1.super_25 534156 114 - 827797 UGAGGCACUGCUUCAGGAACUGUUUGCGCAGCACGCGCAAGUUGUCUAUGGUGUAGCCUGGGGAUCGCAGAU-AUCAGUGGAUUAGUAGAGUCACAGGUCAUAUAAUCUAAACUA ...(((.(((((......((((.((((((.....))))))((.((((..((.....))...)))).))....-..)))).....))))).)))..((((......))))...... ( -29.20, z-score = 0.91, R) >droSim1.chr3R 21524715 114 - 27517382 UGAGGCACUGCUUCAGGAACUGUUUGCGCAGCACGCGCAAGUUGUCUAUGGUGUAGCCUGGGGAUCGCAGAU-AUCAGUGGAUUAGUGGAGUCACCGGUCAUAUAAUCUAAACUA ((((((...))))))(((((((.((((((.....))))))((.((((..((.....))...)))).))....-..)))).(((..(((....)))..)))......)))...... ( -31.40, z-score = 0.66, R) >droMoj3.scaffold_6540 8648357 87 - 34148556 UCAGGCACUGUUUAAGGAAUUGCUUGCGCAGCACGCGCAAGUUAUCGGCGGUAUAGCCUGCAGAGAUAAUAA-AUAAAUAUGUGAACU--------------------------- .....(((((((((.......((((((((.....))))))))((((.((((......))))...))))....-.)))))).)))....--------------------------- ( -25.20, z-score = -1.34, R) >droVir3.scaffold_12855 7115818 90 + 10161210 UCAGGCACUGCUUGAGGAAUUGCUUGCGCAGCACGCGCAAAUUGUCGGCGGUAUAGCCUGCAGGAGAAAUAUUAAAAAUAUAUACAUAUA------------------------- .(((((((((((.((.((.((((..(((.....))))))).)).))))))))...)))))..............................------------------------- ( -23.80, z-score = -0.58, R) >droWil1.scaffold_181108 372783 85 - 4707319 UGAGGCACUGCUUGAGGAAUUGUUUGCGUAAGACGCGCAAAUUGUCCAUGGUGUAGGCUAGAAUAGAAACGU---UAAUACGUGAAAU--------------------------- ...(((.((((....(((...((((((((.....))))))))..))).....))))))).........((((---....)))).....--------------------------- ( -23.50, z-score = -2.02, R) >consensus UGAGGCACUGCUUCAGGAACUGUUUGCGCAGCACGCGCAAGUUGUCGAUGGUGUAGCCUGCAGAACGCAGAU_AACAAUAUAUGAAUUUA_________________________ ..((((..((((....((...((((((((.....))))))))..))...))))..))))........................................................ (-17.28 = -16.52 + -0.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:15 2011