| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,463,518 – 15,463,610 |
| Length | 92 |
| Max. P | 0.932629 |

| Location | 15,463,518 – 15,463,610 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 62.39 |
| Shannon entropy | 0.80128 |
| G+C content | 0.31629 |
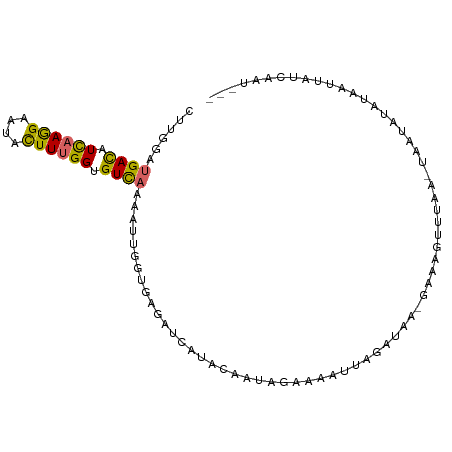
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -6.51 |
| Energy contribution | -6.24 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932629 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
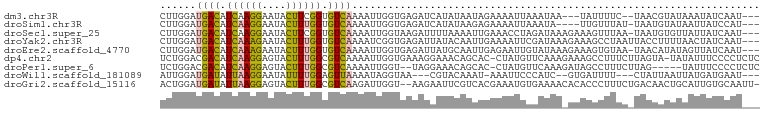
>dm3.chr3R 15463518 92 - 27905053 CUUGGAUGACAUCAAGGAAUACUUCGGUGUCAAAAUUGGUGAGAUCAUAUAAUAGAAAAUUAAAUAA---UAUUUUC--UAACGUAUAAAUAUCAAU--- ......((((((((((.....))).)))))))..(((((((.....((((..((((((((.......---.))))))--))..))))...)))))))--- ( -19.30, z-score = -2.85, R) >droSim1.chr3R 21509535 92 + 27517382 CUUGGAUGACAUCAAGGAAUACUUUGGUGUCAAAAUUGGUGAGAUCAUAUAAGAGAAAAUUAAAUA----UUGUUUAU-UAAUGUAUAAUUAUCCAU--- ..(((((((((((((((....)))))))))).......(((....))).........(((((.(((----(((.....-)))))).)))))))))).--- ( -21.60, z-score = -2.72, R) >droSec1.super_25 513392 96 + 827797 CUUGGAUGACAUCAAGGAAUACUUCGGUGUCAAAAUUGGUAAGAUUUUAAAAUUGAAACCUAGAUAAAGAAAGUUUAA-UAAUGUGUUAUUAUCAAU--- ......((((((((((.....))).)))))))..((((((((.........((((((..((......))....)))))-).........))))))))--- ( -15.77, z-score = -0.76, R) >droYak2.chr3R 4234314 97 - 28832112 CUUGGAUGACAUCAAAGAAUACUUUGGUGUCAAAAUCGGUGAGAUUAUACAAUUGAAAAUUCGAUAAAGAAAGCCUAAUUACCUUUUAACUAUCAAU--- .((((.(((((((((((....))))))))))).....(((((.........(((((....)))))..((.....))..))))).........)))).--- ( -19.70, z-score = -2.30, R) >droEre2.scaffold_4770 11568854 96 + 17746568 CUUGGAUGACAUCAAAGAAUACUUUGGUGUCAAAAUUGGUGAGAUUAUGCAAUUGAGAAUUGUAUAAAGAAAGUGUAA-UAACAUAUAGUUAUCAAU--- ......(((((((((((....)))))))))))..((((((((..(((((((((.....))))))))).....((((..-..))))....))))))))--- ( -26.10, z-score = -4.50, R) >dp4.chr2 29972802 98 - 30794189 UCUGGACGACAUCAAGGAGUACUUUGGCGUCAAAAUUGGUGAAAGGAAACAGCAC-CUAUGUUCAAAGAAAGCCUUUCUUAGUA-UAUAUUUCCCCUCUC ..((((((((..(((((....)))))..)))......((((...(....)..)))-)...)))))(((((.....)))))....-............... ( -21.90, z-score = -0.73, R) >droPer1.super_6 5342036 92 - 6141320 UCUGGACGACAUCAAGGAGUACUUUGGCGUCAAAAUUGGU--UAGGAAACAGCAC-CUAUGUUCAAAGAUAGCCUUUCUUAG-----UAUUUCCCCUCUC ..((((((((..(((((....)))))..)))......(((--..(....)...))-)...)))))((((.......))))..-----............. ( -17.60, z-score = 0.43, R) >droWil1.scaffold_181089 4030615 88 + 12369635 AUUGGAUGAUAUUAAGGAAUAUUUUGGAGUUAAAAUAGGUAA---CGUACAAAU-AAAUUCCCAUC--GUGAUUUU---CUAUUAAUUAUGAUGAAU--- ...............(((((..((((..((((.......)))---)...)))).-..)))))((((--((((((..---.....))))))))))...--- ( -17.30, z-score = -1.56, R) >droGri2.scaffold_15116 843873 97 + 1808639 ACUGGAUGAUAUUAAGGAGUACUUUGGCGUCAAGAUUGGU--AAGAAUUCGUCACGAAAUGUGAAAACACACCCUUUCUGACAACUGCAUUGUGCAAUU- ((..(.(((..((((((....))))))..)))...)..))--........((((.((((((((....))))...))))))))...(((.....)))...- ( -17.80, z-score = 0.53, R) >consensus CUUGGAUGACAUCAAGGAAUACUUUGGUGUCAAAAUUGGUGAGAUCAUACAAUAGAAAAUUAGAUAA_GAAAGUUUAA_UAAUAUAUAAUUAUCAAU___ ......((((.((((((....)))))).)))).................................................................... ( -6.51 = -6.24 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:14 2011