
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,458,567 – 15,458,733 |
| Length | 166 |
| Max. P | 0.981385 |

| Location | 15,458,567 – 15,458,659 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 87.79 |
| Shannon entropy | 0.21724 |
| G+C content | 0.50917 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
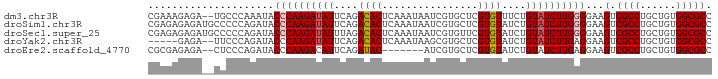
>dm3.chr3R 15458567 92 + 27905053 CGAAAGAGA--UGCCCAAAUACCCAAGAUAUUCAGACACUCAAAUAAUCGUGCUCGUGUUUCUGUAUCUUGGGGAAGUCGCCUGCUGUGGCGCC (....)...--..........((((((((((..((((((................))))))..))))))))))...(.((((......))))). ( -27.59, z-score = -2.13, R) >droSim1.chr3R 21504570 94 - 27517382 CGAGAGAGAUGCCCCCAGAUACCCAAGAUAUUCAGACACUCAAAUAAUCGUGCUCGUGUAUCUGUAUCUUGGGGAAGUCGCCUGCUGUGGCGCC .......(((..((((((((((...((((((...(((((..........))).))..))))))))))).)))))..)))(((......)))... ( -30.90, z-score = -2.25, R) >droSec1.super_25 508443 94 - 827797 CGAGAGAGAUGCCCCCAGAUACCCAAGAUAUUUAGACACUCAAAUAAUCGUGUUCGUGUAUCUGUAUCUUGGGGAAGUCGCCUGCUGUGGCGCC .......(((..((((((((((...((((((...(((((..........)))))...))))))))))).)))))..)))(((......)))... ( -32.10, z-score = -3.16, R) >droYak2.chr3R 4229537 87 + 28832112 -----GAGA--UUCCCAGAUACCCAAGAUAUUCAGACACUCAAAUAAGCGUGCUCGUGUAUCUGUAUCUUGAGGAAGUCGCCUGCUGUGGCGCC -----((..--(((((((((((...((((((...(((((.(......).))).))..))))))))))).)).)))).))(((......)))... ( -25.20, z-score = -1.87, R) >droEre2.scaffold_4770 11563327 85 - 17746568 CGCGAGAGA--CUCCCAGAUACCCAAGACAUUCAGAUAC-------AUCGUGCUCGUGUAUCUGUAUCUUGAGGAAGUCGCCUGCUGUGGCGCC .((....((--((.((.(....)(((((....(((((((-------(.((....))))))))))..))))).)).))))(((......))))). ( -28.10, z-score = -1.91, R) >consensus CGAGAGAGA__CCCCCAGAUACCCAAGAUAUUCAGACACUCAAAUAAUCGUGCUCGUGUAUCUGUAUCUUGGGGAAGUCGCCUGCUGUGGCGCC .......((..((((.((((((...((((((...(((((..........))).))..)))))))))))).))))...))(((......)))... (-20.84 = -21.48 + 0.64)

| Location | 15,458,567 – 15,458,659 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 87.79 |
| Shannon entropy | 0.21724 |
| G+C content | 0.50917 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
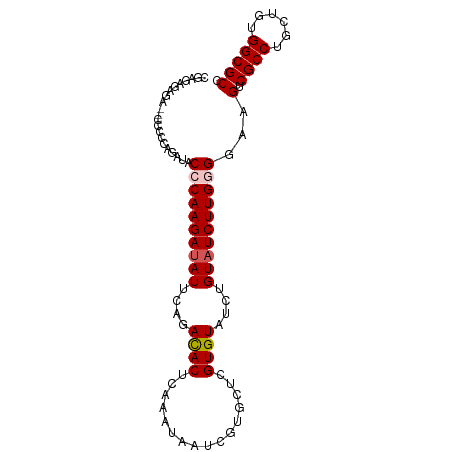
>dm3.chr3R 15458567 92 - 27905053 GGCGCCACAGCAGGCGACUUCCCCAAGAUACAGAAACACGAGCACGAUUAUUUGAGUGUCUGAAUAUCUUGGGUAUUUGGGCA--UCUCUUUCG .(((((......)))......(((((((((((((.((.((((........)))).)).))))..))))))))).......)).--......... ( -25.60, z-score = -1.41, R) >droSim1.chr3R 21504570 94 + 27517382 GGCGCCACAGCAGGCGACUUCCCCAAGAUACAGAUACACGAGCACGAUUAUUUGAGUGUCUGAAUAUCUUGGGUAUCUGGGGGCAUCUCUCUCG ..((((......)))).....((((((((((((((((.((((........)))).)))))))..))))))))).....(((((.....))))). ( -33.00, z-score = -2.60, R) >droSec1.super_25 508443 94 + 827797 GGCGCCACAGCAGGCGACUUCCCCAAGAUACAGAUACACGAACACGAUUAUUUGAGUGUCUAAAUAUCUUGGGUAUCUGGGGGCAUCUCUCUCG ..((((......)))).....(((((((((.((((((.((((........)))).))))))...))))))))).....(((((.....))))). ( -30.40, z-score = -2.75, R) >droYak2.chr3R 4229537 87 - 28832112 GGCGCCACAGCAGGCGACUUCCUCAAGAUACAGAUACACGAGCACGCUUAUUUGAGUGUCUGAAUAUCUUGGGUAUCUGGGAA--UCUC----- ..((((......)))).....((((((((((((((((.((((........)))).)))))))..)))))))))..........--....----- ( -26.20, z-score = -1.70, R) >droEre2.scaffold_4770 11563327 85 + 17746568 GGCGCCACAGCAGGCGACUUCCUCAAGAUACAGAUACACGAGCACGAU-------GUAUCUGAAUGUCUUGGGUAUCUGGGAG--UCUCUCGCG .(((((......)))(((((((((((((((((((((((((....)).)-------)))))))..))))))))......)))))--))....)). ( -34.40, z-score = -3.46, R) >consensus GGCGCCACAGCAGGCGACUUCCCCAAGAUACAGAUACACGAGCACGAUUAUUUGAGUGUCUGAAUAUCUUGGGUAUCUGGGAG__UCUCUCUCG ..((((......)))).....((((((((((((((((.((((........)))).)))))))..))))))))).....(((.....)))..... (-26.06 = -26.62 + 0.56)


| Location | 15,458,635 – 15,458,733 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 70.79 |
| Shannon entropy | 0.52068 |
| G+C content | 0.59306 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.39 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642033 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
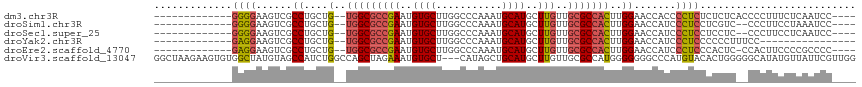
>dm3.chr3R 15458635 98 + 27905053 -------------GGGGAAGUCGCCUGCUG--UGGCGCCGAAUGUGCUUGGCCCAAAUGCAUGCUUGUUGCGCCACUUGGAACCACCCCUCUCUCUCACCCCUUUCUCAAUCC---- -------------((((..((..((....(--(((((((((..((((...........))))..)))..)))))))..)).))..))))........................---- ( -29.20, z-score = -1.11, R) >droSim1.chr3R 21504640 96 - 27517382 -------------GGGGAAGUCGCCUGCUG--UGGCGCCGAAUGUGCUUGGCCCAAAUGCAUGCUUGUUGCGCCACUUGGAACCAUCCCUCCUCGUC--CCCUUCCUAAAUCC---- -------------(((((...........(--(((((((((..((((...........))))..)))..)))))))..(((........)))...))--)))...........---- ( -30.30, z-score = -1.58, R) >droSec1.super_25 508513 96 - 827797 -------------GGGGAAGUCGCCUGCUG--UGGCGCCGAAUGUGCUUGGCCCAAAUGCAUGCUUGUUGCGCCACUUGGAACCAUCCCUCCUCCUC--CCCUUCCUCAAUCC---- -------------(((((((.........(--(((((((((..((((...........))))..)))..)))))))..(((........))).....--..))))))).....---- ( -31.20, z-score = -2.00, R) >droYak2.chr3R 4229600 86 + 28832112 -------------GAGGAAGUCGCCUGCUG--UGGCGCCGAAUGUGCUUGGCCCAAAUGCAUGCUUGUUGCGCCACUUGGAACCAUCCCUCCCCCCUUUCC---------------- -------------((((......((....(--(((((((((..((((...........))))..)))..)))))))..)).......))))..........---------------- ( -26.02, z-score = -1.29, R) >droEre2.scaffold_4770 11563388 97 - 17746568 -------------GAGGAAGUCGCCUGCUG--UGGCGCCGAAUGUGCUUGGCCCAAAUGCAUGCUUGUUGCGCCACUUGGAACCAUCCCUCCCACUC-CCACUUCCCCGCCCC---- -------------(.((((((........(--(((((((((..((((...........))))..)))..)))))))..(((........))).....-..)))))).).....---- ( -28.10, z-score = -1.20, R) >droVir3.scaffold_13047 3102605 114 - 19223366 GGCUAAGAAGUGUGGCUAUGUAGCCAUCUGGCCAGCUAGAAAUGUGCU---CAUAGCUGCAUGCUUGUUGCGCCAUGGGGGGGCCCAUGUACACUGGGGGCAUAUGUUAUUCGUUGG ((((((.(((((..((((((.((((((((((....)))))..)).)))---))))))..)..)))).))).)))((((.(..((((..(....)...))))...).))))....... ( -39.80, z-score = 0.61, R) >consensus _____________GGGGAAGUCGCCUGCUG__UGGCGCCGAAUGUGCUUGGCCCAAAUGCAUGCUUGUUGCGCCACUUGGAACCAUCCCUCCCCCUC__CCCUUCCUCAAUCC____ .............((((......((.......(((((((((..((((...........))))..)))..))))))...)).......)))).......................... (-16.86 = -17.39 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:14 2011