
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,452,041 – 15,452,136 |
| Length | 95 |
| Max. P | 0.697911 |

| Location | 15,452,041 – 15,452,136 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.52 |
| Shannon entropy | 0.49529 |
| G+C content | 0.43980 |
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -13.24 |
| Energy contribution | -12.91 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15452041 95 - 27905053 -------------GAGAACCAUUAUCUCGGGCCAACCAGUUGGAUCUGGUAUGGAG--------AUACAUAUUGGCGCGA----UUAUCUUGAUUGCAUUCGUGGAAGCGGUCUCCUUAG -------------((((.((.((.((.((((((((((((......))))((((...--------...)))))))))((((----((.....))))))..))).)).)).))))))..... ( -27.80, z-score = -1.00, R) >droSim1.chr3R 21497433 95 + 27517382 -------------GAGAACCAUUAUCUCGGGCCAACCAGUUGGAUCUGGUAUGGAG--------AUACAUAUUGGCGCGA----UUAUCUUGAUUGCAUUCGUGGAAGCGGUCUCCUUAG -------------((((.((.((.((.((((((((((((......))))((((...--------...)))))))))((((----((.....))))))..))).)).)).))))))..... ( -27.80, z-score = -1.00, R) >droSec1.super_25 502689 95 + 827797 -------------GAGAACCAUUAUCUCGGGCCAACCAGUUGGAUCUGGUAUGGAG--------AUACAUAUUGGCGCGA----UUAUCUUGAUUGCAUUCGUGGAAGCGGUCUCCUUAG -------------((((.((.((.((.((((((((((((......))))((((...--------...)))))))))((((----((.....))))))..))).)).)).))))))..... ( -27.80, z-score = -1.00, R) >droYak2.chr3R 4222972 95 - 28832112 -------------GAGAACCAUUAUCUCGGGGCAACCAGUUGGAUCUGGUAUGAUG--------AUACAUAUUGGCACGA----UAAUCUUGAUUGCAUUCGUGGAAGCAGUCUCCUUAG -------------((((.......)))).((....)).((((...(..(((((...--------...)))))..)..)))----)......((((((.((....)).))))))....... ( -23.20, z-score = -0.17, R) >droEre2.scaffold_4770 11556875 88 + 17746568 --------------------AUUAUCUCGGGGCAACCAGUUGGAUCUGGUAUGGAG--------AUACACAUUUCCACGA----UAAUCUUGAUUGCAUUCGUGGAAGCAGUUUCCUUGG --------------------..((((((.(....(((((......))))).).)))--------)))....(((((((((----((((....))))...)))))))))............ ( -23.70, z-score = -1.04, R) >droAna3.scaffold_13340 8748509 93 + 23697760 -------------AAGAACUAUUAUCUCUGG-CAACCAGUUGAAUCUGGUAUCGGA--------AUAUAUAUUCGCACAG----UAAUCUUGAUUGGUUUCGUGGAAGCAAUCGCAUAG- -------------.....((((.....((((-(.(((((......)))))....((--------((....)))))).)))----.......(((((.(((....))).)))))..))))- ( -20.30, z-score = -0.93, R) >dp4.chr2 29961607 120 - 30794189 GGGAACCAUUAUCUUGGACGUCUACGACUGGGGAACCAGUUGGAUCUGGUAUAUUAUUUUAUAUGCAUAUACUCGUACAGAUGAUAAUCUCGAUUGCUUUCGUGGAAGCAAUCUCCUUGG ((((...(((((((...(((..(((((((((....)))))))......((((((......))))))...))..)))..)))))))..))))(((((((((....)))))))))....... ( -39.60, z-score = -3.78, R) >droPer1.super_6 5330866 120 - 6141320 GGGAACUAUUAUCUUGGACGUCUACGACUGGGGAACCAGUUGGAUCUGGUAUAUUAUUUUAUAUGCAUAUACUCGUACAGAUGAUAAUCUCGAUUGCUUUCGUGGAAGCAAUCUCCUUGG ((((..((((((((...(((..(((((((((....)))))))......((((((......))))))...))..)))..)))))))).))))(((((((((....)))))))))....... ( -40.90, z-score = -4.55, R) >consensus _____________GAGAACCAUUAUCUCGGGGCAACCAGUUGGAUCUGGUAUGGAG________AUACAUAUUGGCACGA____UAAUCUUGAUUGCAUUCGUGGAAGCAGUCUCCUUAG .............................((...(((((......))))).........................................((((((.((....)).)))))).)).... (-13.24 = -12.91 + -0.33)

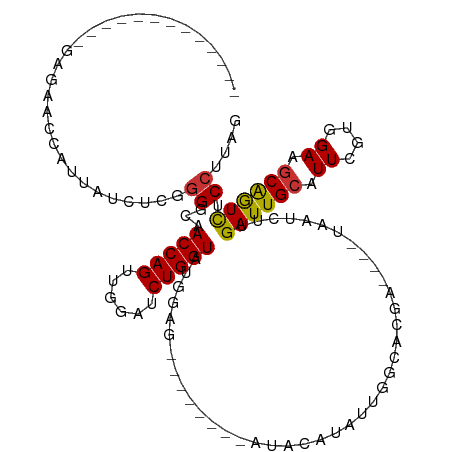
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:08 2011