
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,035,191 – 8,035,321 |
| Length | 130 |
| Max. P | 0.872954 |

| Location | 8,035,191 – 8,035,287 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.66 |
| Shannon entropy | 0.55415 |
| G+C content | 0.54513 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -17.70 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
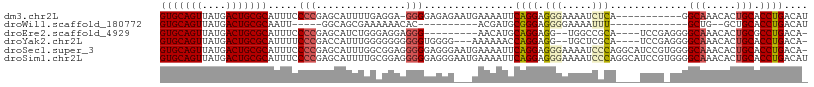
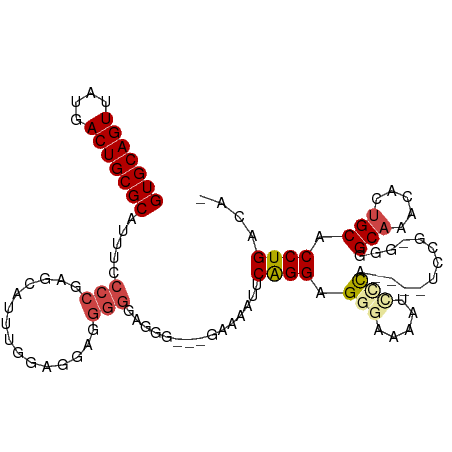
>dm3.chr2L 8035191 96 - 23011544 GUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUUUUGAGGA-GGGGAGAGAAUGAAAAUUCAGGAGGGAAAAUCUCA-----------GGCAAACACUGCACCUGACAU (((((((....))))))).(((((((...(.....)....-)))))))..........(((((.(((.....))).-----------.(((.....))).)))))... ( -32.00, z-score = -2.74, R) >droWil1.scaffold_180772 5388545 78 + 8906247 GUGCAGUUAUGACUGCGCAAUU-----GGCAGCGAAAAAACAC----------ACGAUGCGGGAGGGGAAAAUUU-------------GCUG--GCUGCACCUGACAU (((((((....)))))))....-----.((((((......)..----------......(((.(((......)))-------------.)))--)))))......... ( -20.40, z-score = -0.38, R) >droEre2.scaffold_4929 16940738 92 - 26641161 GUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUCUGGGAGGAGGG---------AACAUGCAGGAGG--UGGCCGCA----UCCGAGGGGCAAACACUGCGCCUGACA- (((((((....))))))).(((((((.((...)).)).))))).---------....((((((.((--(((((.(.----.....).)))...))))...)))).))- ( -33.40, z-score = -0.31, R) >droYak2.chr2L 10664869 98 - 22324452 GUGCAGUUAUGACUGCGCAUUUUCCCGACCAUUUGGGGGGGGGGUGGGG---AAAAAACCAGGAGG--UGCUCGCA----UCCGAGGGGCAAACACUGCACCUGACA- (((((((....))))))).((..(((((....)))))..))((((((((---.....(((....))--).))).))----))).(((.(((.....))).)))....- ( -36.80, z-score = -1.08, R) >droSec1.super_3 3532059 107 - 7220098 GUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUUUGGCGGAGGGGGAGGGAAUGAAAAUUCAGGAGGGAAAAUCCCAGGCAUCCGUGGGGCAAACACUGCACCUGACA- (((((((....))))))).(((((((..((.....))...))))))).((((....)))).(..(((.....((((.(....).))))(((.....))).)))..).- ( -40.20, z-score = -1.79, R) >droSim1.chr2L 7815348 108 - 22036055 GUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUUUUGCGGAGGGGGAGGGAAUGAAAAUUCAGGAGGGAAAAUCCCAGGCAUCCGUGGGGCAAACACUGCACCUGACAU (((((((....))))))).(((((((..((.....))...))))))).((((....)))).(..(((.....((((.(....).))))(((.....))).)))..).. ( -40.60, z-score = -2.12, R) >consensus GUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUUUGGAGGAGGGGGAGGG___GAAAAUUCAGGAGGGAAAAUCCCA____UCCG_GGGGCAAACACUGCACCUGACA_ (((((((....))))))).....(((...............)))...............((((.(((.....))).............(((.....))).)))).... (-17.70 = -18.44 + 0.74)
| Location | 8,035,218 – 8,035,321 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.73 |
| Shannon entropy | 0.69581 |
| G+C content | 0.48861 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -8.56 |
| Energy contribution | -8.18 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
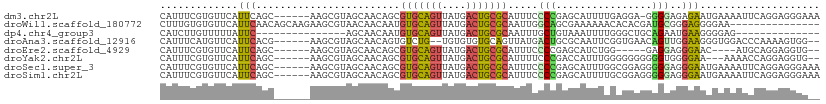
>dm3.chr2L 8035218 103 - 23011544 CAUUUCGUGUUCAUUCAGC------AAGCGUAGCAACAGCGUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUUUUGAGGA-GGGGAGAGAAUGAAAAUUCAGGAGGGAAA ..((((.(((((((((.((------..((((.......))))(((((....))))))).(((((((...(.....)....-))))))))))))))....)).)))).... ( -32.30, z-score = -2.38, R) >droWil1.scaffold_180772 5388568 95 + 8906247 CUUUGUGUGUUCAUUCAACAGCAAGAAGCGUAACAACAAUGUGCAGUUAUGACUGCGCAAUUGGCAGCGAAAAAACACACGAUGCGGGAGGGGAA--------------- ..(((((((((..(((....((.....)).......(((((((((((....))))))).)))).....)))..))))))))).............--------------- ( -27.10, z-score = -1.31, R) >dp4.chr4_group3 7857803 81 + 11692001 CAUCUUGUUUUUAUUC---------------AGCAACAAUGUGCAGUUAUGACUGCGCAAUUUGCUGUAAAUUUUGGGCUGCAGAAUGAAGGGGAG-------------- ...(((.(((((((((---------------(((((...((((((((....))))))))..)))))(((..........))).))))))))).)))-------------- ( -25.10, z-score = -2.40, R) >droAna3.scaffold_12916 9525667 100 + 16180835 CAUUUCAUGUUCAUUCACG------AAGCGUAGCAACAGUGUCUG--UGUGUGUGCAGUUAUGACUGCGCAAUUCGGUGAACAGUUGGAAGGGUGGACCCAAAAGUGG-- ..((((((((((((...((------((((((((((....)).)))--))).((((((((....)))))))).)))))))))))..)))))(((....)))........-- ( -30.30, z-score = -1.03, R) >droEre2.scaffold_4929 16940771 93 - 26641161 CAUUUCGUGUUCAUUCAGC------AAGCGUAGCAACAGCGUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUCUGG-----GAGGAGGGAAC----AUGCAGGAGGUG-- ((((((.(((...(((.((------..((((.......))))(((((....))))))).(((((((.((...)).)-----).)))))))).----..))).))))))-- ( -29.50, z-score = -0.96, R) >droYak2.chr2L 10664902 99 - 22324452 CAUUUCGUGUUCAUUCAGC------AAGCGUAGCAACAGCGUGCAGUUAUGACUGCGCAUUUUCCCGACCAUUUGGGGGGGGGGUGGGGAA---AAAACCAGGAGGUG-- ((((((.((........((------..((((.......))))(((((....))))))).(((((((.(((............))).)))))---))...)).))))))-- ( -31.50, z-score = -1.12, R) >droSec1.super_3 3532096 104 - 7220098 CAUUUCGUGUUCAUUCAGC------AAGCGUAGCAACAGCGUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUUUGGCGGAGGGGGAGGGAAUGAAAAUUCAGGAGGGAAA ..((((.(((((((((.((------..((((.......))))(((((....))))))).(((((((..((.....))...))))))).)))))))....)).)))).... ( -34.60, z-score = -2.22, R) >droSim1.chr2L 7815386 104 - 22036055 CAUUUCGUGUUCAUUCAGC------AAGCGUAGCAACAGCGUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUUUUGCGGAGGGGGAGGGAAUGAAAAUUCAGGAGGGAAA ..((((.(((((((((.((------..((((.......))))(((((....))))))).(((((((..((.....))...))))))).)))))))....)).)))).... ( -35.00, z-score = -2.44, R) >consensus CAUUUCGUGUUCAUUCAGC______AAGCGUAGCAACAGCGUGCAGUUAUGACUGCGCAUUUCCCCGAGCAUUUGGGGGAGGGGGAGGGAAGGAAAAUUCAGGAGGGA__ .............(((........................(((((((....))))))).....(((................)))..))).................... ( -8.56 = -8.18 + -0.39)
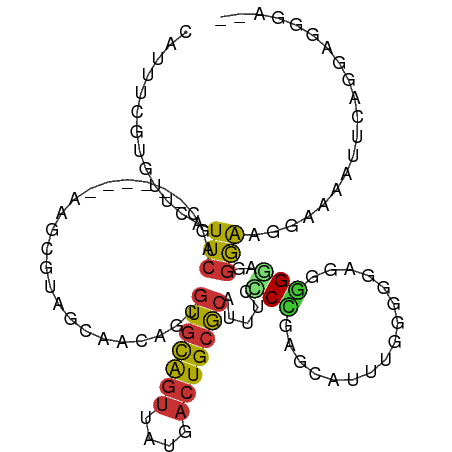

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:55 2011