
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,444,799 – 15,444,900 |
| Length | 101 |
| Max. P | 0.543528 |

| Location | 15,444,799 – 15,444,900 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Shannon entropy | 0.42075 |
| G+C content | 0.42237 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -13.12 |
| Energy contribution | -12.90 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
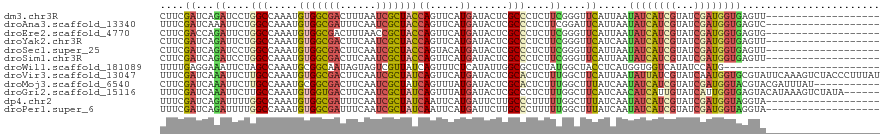
>dm3.chr3R 15444799 101 - 27905053 CUUCGAUCAGAUCCUGGCCAAAUGUGGCGACUUUAAUCGCUACCAGUUCAUGAUACUCGCCCUCUUCGGGUUCAUUAAUAUCAUCGUAUCGAUGGUGAGUU------------------- .....((((((..((((.......((((((......)))))))))).)).))))((((((((.....))))........(((((((...))))))))))).------------------- ( -26.81, z-score = -1.31, R) >droAna3.scaffold_13340 8741767 101 + 23697760 UUUCGAUCAAAUUCUGGCCAAAUGUGGCGAUUUCAAUCGCUACCAGUUCAUGAUACUCGCCCUCUUCGGAUUCAUUAAUAUCAUCGUAUCGAUGGUGAGUC------------------- .....((((....((((.......(((((((....)))))))))))....))))(((((((.((....))............((((...))))))))))).------------------- ( -25.81, z-score = -1.67, R) >droEre2.scaffold_4770 11549455 101 + 17746568 CUUCGACCAGAUUCUGGCCAAAUGUGGCGACUUUAACCGCUACCAGUUCAUGAUACUCGCCCUCUUCGGGUUCAUUAAUAUCAUCGUAUCGAUGGUGAGUG------------------- ....(.((((...)))).)....((((((........))))))..........(((((((((.....))))........(((((((...))))))))))))------------------- ( -26.20, z-score = -1.15, R) >droYak2.chr3R 4215575 101 - 28832112 CUUCGAUCAGAUUCUGGCCAAAUGUGGCGACUUCAAUCGCUACCAGUUCAUGAUACUCGCCCUCUUCGGGUUCAUCAAUAUCAUCGUAUCGAUGGUGAGUU------------------- .....((((....((((.......((((((......))))))))))....))))((((((((.....))))........(((((((...))))))))))).------------------- ( -27.41, z-score = -1.37, R) >droSec1.super_25 495601 101 + 827797 CUUCGAUCAGAUCCUGGCCAAAUGUGGCGACUUCAAUCGCUACCAGUACAUGAUACUCGCCCUCUUCGGGUUCAUUAAUAUCAUCGUAUCGAUGGUGAGUU------------------- .............((.((((...(((((((......)))))))(.((((((((((...((((.....)))).......)))))).)))).).)))).))..------------------- ( -26.80, z-score = -1.18, R) >droSim1.chr3R 21490329 101 + 27517382 CUUCGAUCAGAUCCUGGCCAAAUGUGGCGACUUCAAUCGCUACCAGUUCAUGAUACUCGCCCUCUUCGGGUUCAUUAAUAUCAUCGUAUCGAUGGUGAGUU------------------- .....((((((..((((.......((((((......)))))))))).)).))))((((((((.....))))........(((((((...))))))))))).------------------- ( -26.81, z-score = -1.20, R) >droWil1.scaffold_181089 2611568 94 - 12369635 UUUUGAGGAAAUUCUAGCCAAAUGCGGCAAUAGUAGUCGUUAUCAGUUUCUCAUAUUGGCGCUCUAUGGCUACCUCAUGGUGGUCAUAUCCAUG-------------------------- ......(((.......(((((..(((((.......))))).....(.....)...)))))....((((((((((....)))))))))))))...-------------------------- ( -27.00, z-score = -2.36, R) >droVir3.scaffold_13047 3083427 120 + 19223366 UUUCGAUCAAAUUCUUGCCAAAUGUGGCGACUUCAAUCGCUAUCAGUUCAUGAUACUCGCACUCUUUGGCUUCAUUAAUAUUAUCGUAUCAAUGGUGCGUAUUCAAAGUCUACCCUUUAU ....(((.........((((((.(((((((......))))(((((.....)))))....)))..))))))..............(((((.....)))))........))).......... ( -21.80, z-score = -1.39, R) >droMoj3.scaffold_6540 19204596 109 + 34148556 CUUCGAUCAAAUUCUUGCCAAAUGCGGCGACUUCAAUCGCUAUCAGUUUAUGAUACUCGCACUCUUUGGCUUUAUCAAUAUCAUCGUAUCGAUGGUACGUACGAUUUAU----------- ..(((...........((((((((((((((......))))(((((.....)))))..))))...))))))........((((((((...))))))))....))).....----------- ( -27.80, z-score = -3.05, R) >droGri2.scaffold_15116 812857 114 + 1808639 UUUCGAUCAAAUUCUUGCCAAAUGUGGUGACUUCAAUCGCUAUCAGUUUAUGAUACUCGCCCUCUUUGGCUUCAUCAACAUCAUUGUAUCAUUGGUGAGUACAUAAAGUCUAUA------ ....(((......((..(((((((..((((..........(((((.....)))))...(((......)))..........))))..)))..))))..))........)))....------ ( -22.24, z-score = -1.06, R) >dp4.chr2 29952650 101 - 30794189 UUUCGAUCAGAUUUUGGCCAAAUGUGGCGAUUUCAAUCGCUAUCAAUUCAUGAUUCUUGCCCUUUUUGGCUUUAUCAAUAUCAUCGUAUCGAUGGUAGGUA------------------- .....(((.(((...(((((((..(((((((....)))))))(((.....)))...........)))))))..)))..((((((((...))))))))))).------------------- ( -26.40, z-score = -2.15, R) >droPer1.super_6 5321880 101 - 6141320 UUUCGAUCAGAUUUUGGCCAAAUGUGGCGAUUUCAAUCGCUAUCAAUUCAUGAUUCUUGCCCUUUUUGGCUUUAUCAAUAUCAUCGUAUCGAUGGUAGGUA------------------- .....(((.(((...(((((((..(((((((....)))))))(((.....)))...........)))))))..)))..((((((((...))))))))))).------------------- ( -26.40, z-score = -2.15, R) >consensus CUUCGAUCAGAUUCUGGCCAAAUGUGGCGACUUCAAUCGCUACCAGUUCAUGAUACUCGCCCUCUUCGGCUUCAUCAAUAUCAUCGUAUCGAUGGUGAGUA___________________ ....((...((....(((.....(((((((......)))))))((.....))......)))....))....)).....((((((((...))))))))....................... (-13.12 = -12.90 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:05 2011