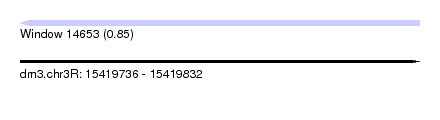
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,419,736 – 15,419,832 |
| Length | 96 |
| Max. P | 0.845332 |

| Location | 15,419,736 – 15,419,832 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Shannon entropy | 0.47488 |
| G+C content | 0.59068 |
| Mean single sequence MFE | -37.01 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
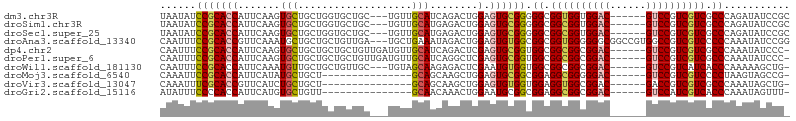
>dm3.chr3R 15419736 96 - 27905053 UAAUAUCCGCACCAUUCAAGUGCUGCUGGUGCUGC---UGUUGCAUCAGACUGGAGUGCGGGGGCGGUGGUGGAC------GUCCGUCGUCGCCCAGAUAUCCGC ......((((((..((((.((....(((((((...---....)))))))))))))))))))(((((..(..((..------..))..)..))))).......... ( -40.20, z-score = -1.43, R) >droSim1.chr3R 21464763 96 + 27517382 UAAUAUCCGCACCAUUCAAGUGCUGCUGGUGCUGC---UGUUGCAUGAGACUGGAGUGCGGGGGCGGCGGUGGAC------GUCCGUCGUCGCCCAGAUAUCCGC ......((((((((.((..((((.((.((.....)---))).))))..)).)))..)))))((((((((..((..------..))..)))))))).......... ( -41.70, z-score = -1.56, R) >droSec1.super_25 470695 96 + 827797 UAAUAUCCGCACCAUUCAAGUGCUGCUGGUGCUGC---UGUUGCAUGAGACUGGAGUGCGGGGGCGGCGGUGGAC------GUCCGUCGUCGCCCAGAUAUCCGC ......((((((((.((..((((.((.((.....)---))).))))..)).)))..)))))((((((((..((..------..))..)))))))).......... ( -41.70, z-score = -1.56, R) >droAna3.scaffold_13340 8716362 102 + 23697760 CAAUUUCCGCACCGUUCAAAUGCUGCUGCUGUUGA---UGCUGAAAUAGACUGGAGUGUGGCGGCGGUGGGGGGCGGCCGUGGCCGUCGUCCCCCAAAUAUCCGG ......((((((((((...(((((.(..(((((..---......)))))...).)))))...)))))))(((((((((....))))....))))).......))) ( -39.20, z-score = -0.46, R) >dp4.chr2 29929756 98 - 30794189 CAAUUUCCGCACCAUUCAAGUGCUGCUGCUGCUGUUGAUGUUGCAUCAGACUCGAGUGCGGUGGCGGCGGCGGAC------GUCCGUCGUCGCCCAAAUAUCCC- ......((((((...((.(((.(((.(((.((.......)).))).)))))).)))))))).(((((((((((..------..)))))))))))..........- ( -43.20, z-score = -3.37, R) >droPer1.super_6 5298901 98 - 6141320 CAAUUUCCGCACCAUUCAAGUGCUGCUGCUGCUGUUGAUGUUGCAUCAGGCUCGAGUGCGGUGGCGGCGGCGGAC------GUCCGUCGUCGCCCAAAUAUCCC- ......((((((...((.(((.(((.(((.((.......)).))).)))))).)))))))).(((((((((((..------..)))))))))))..........- ( -42.60, z-score = -2.70, R) >droWil1.scaffold_181130 4253488 95 - 16660200 CAAUUUCCGCACCAUUCAAAUGUUGCUGCUGUUGC---UGUAGCAAGAGACUCGAAUGUGGUGGCGGCGGCGGAC------GUCCGUCAUCACCCAAAAAGCUG- ........((...((((...(.(((((((......---.))))))).).....))))((((((((((((.....)------).)))))))))).......))..- ( -31.30, z-score = -1.21, R) >droMoj3.scaffold_6540 21664498 83 + 34148556 CAAAUUCCGCACCAUUCAUAUGCUGCU---------------GCAGCAAGCUGGAGUGCGGCGGAGGCGGGGGAC------GUCCGUCGUCCCCUAAGUAGCCG- .....(((((..(((((....(((((.---------------...)).)))..)))))..)))))((((((((((------(.....)))))))).....))).- ( -40.20, z-score = -3.43, R) >droVir3.scaffold_13047 3054925 83 + 19223366 CAAAUUUCGCACCGUUCAUCUGCUGCU---------------GCAGCAAGCUGGAGUGUGGUGGAGGUGGCGGAC------GACCGUCGUCGCCCAAAUAGCUG- ........(((((.((((((.((.(((---------------.(((....))).))))))))))))))((((.((------(.....)))))))......))..- ( -28.20, z-score = -0.17, R) >droGri2.scaffold_15116 769898 83 + 1808639 AUAUUUCCCCACCAUUCAUGUGCUGUU---------------GCAACAAACUGGAAUGCGGCGGAGGCGGCGGAC------GUCCAUCGUCACCCAAAUAGUUU- ...(((((((..(((((.(((((....---------------)).))).....))))).)).))))).((..(((------(.....))))..)).........- ( -21.80, z-score = 0.14, R) >consensus CAAUUUCCGCACCAUUCAAGUGCUGCUGCUGCUGC___UGUUGCAUCAGACUGGAGUGCGGUGGCGGCGGCGGAC______GUCCGUCGUCGCCCAAAUAUCCG_ ......(((((((((....))).(((................)))..........)))))).((.((((((((((......)))))))))).))........... (-20.56 = -20.66 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:28:04 2011