
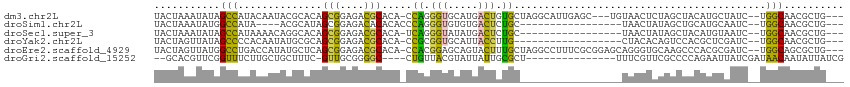
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,998,749 – 7,998,855 |
| Length | 106 |
| Max. P | 0.857934 |

| Location | 7,998,749 – 7,998,855 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 54.40 |
| Shannon entropy | 0.80186 |
| G+C content | 0.50648 |
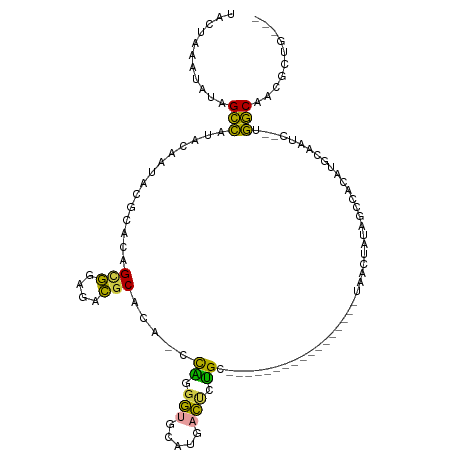
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -9.91 |
| Energy contribution | -8.75 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.82 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7998749 106 - 23011544 UACUAAAUAUAGCCAUACAAUACGCACAGCGGAGACGCACA-CCAGGGUGCAUGACUGUGCUAGGCAUUGAGC---UGUAACUCUAGCUACAUGCUAUC--UGGCAACGCUG--- ...........((((........((((((((....)((((.-.....))))..).))))))..(((((..(((---((......)))))..)))))...--)))).......--- ( -33.90, z-score = -1.73, R) >droSim1.chr2L 7788486 89 - 22036055 UACUAAAUAUGGCCAUA----ACGCAUAGCGGAGACACACACCCAGGGUGUGUGACUCUGC-----------------UAACUAUAGCUGCAUGCAAUC--UGGCAACGCUG--- ...........((((..----.....((((((((.((((((((...)))))))).))))))-----------------))......((.....))....--)))).......--- ( -32.90, z-score = -3.60, R) >droSec1.super_3 3505490 92 - 7220098 UACUAAAUAUAGCCAUAAAACAGGCACAGCGGAGACGCACA-UCAGGGUAUAUGACUCUGC-----------------UAACUAUAGCUACAUGUAAUC--UGGCAACGCUG--- ...........(((........)))...(((....)))...-((((..((((((.....((-----------------((....))))..))))))..)--)))........--- ( -23.50, z-score = -2.01, R) >droYak2.chr2L 10632611 91 - 22324452 UACUAGUUAUAGCCCCACAAUAUGCGCAGCGGAGACGCACA-CCGCGGUGCAUUACCUUG------------------CUACACAGUCCACGCUCGAUC--UGGCAACGCUG--- .........((((........((((((.((((.(.....).-)))).))))))......)------------------)))..((((....(((.....--.)))...))))--- ( -21.44, z-score = 0.63, R) >droEre2.scaffold_4929 16913526 109 - 26641161 UACUAGUUAUGGCCUGACCAUAUGCUCAGCGGAGACGCACA-CCACGGAGCAGUACUUUGCUAGGCCUUUCGCGGAGCAGGGUGCAAGCCCACGCGAUC--UGGCAGCGCUG--- ..........((((((.(((.((((...(((....)))...-....((.((.((((((((((..((.....))..))))))))))..))))..)).)).--)))))).))).--- ( -40.40, z-score = -0.77, R) >droGri2.scaffold_15252 5602400 93 + 17193109 --GCACGUUCGGUUUCUUGCUGCUUUC-GUUGCGGGGC----CUGUUACGUAUUAUUGCGCU---------------UUUCGUUCGCCCCAGAAUUAUCGAUAACAAUAUUAUCG --(((((..((((.....))))....)-).)))(((((----.((...((((....))))..---------------...))...)))))........((((((.....)))))) ( -21.70, z-score = -0.58, R) >consensus UACUAAAUAUAGCCAUACAAUACGCACAGCGGAGACGCACA_CCAGGGUGCAUGACUCUGC_________________UAACUAUAGCCACAUGCAAUC__UGGCAACGCUG___ ...........(((..............(((....))).....((.(((.....))).))..........................................))).......... ( -9.91 = -8.75 + -1.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:54 2011