
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,377,786 – 15,377,876 |
| Length | 90 |
| Max. P | 0.647625 |

| Location | 15,377,786 – 15,377,876 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.73 |
| Shannon entropy | 0.54284 |
| G+C content | 0.52133 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -7.08 |
| Energy contribution | -7.27 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
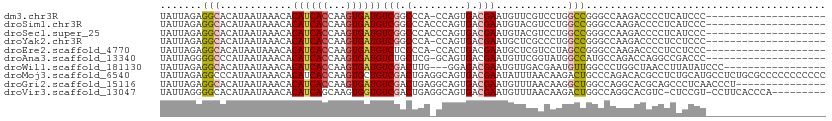
>dm3.chr3R 15377786 90 - 27905053 UAUUAGAGGCACAUAAUAAACACAUCACCAAGUGAUGUCGGCCCA-CCAGUGACGAAUGUUCGUCCUGGCCGGGCCAAGACCCCUCAUCCC-------------------- .....((((............(((((((...))))))).(((((.-((((.(((((....)))))))))..)))))......)))).....-------------------- ( -36.80, z-score = -5.67, R) >droSim1.chr3R 21422068 91 + 27517382 UAUUAGAGGCACAUAAUAAACACAUCACCAAGUGAUGUCGGCCCACCCAGUGACGAAUGUACGUCCUGGCCGGGCCAAGACCCCUCAUCCC-------------------- .....((((............(((((((...))))))).(((((..((((.((((......))))))))..)))))......)))).....-------------------- ( -35.60, z-score = -5.23, R) >droSec1.super_25 429302 91 + 827797 UAUUAGAGGCACAUAAUAAACACAUCACCAAGUGAUGUCGGCCCACCCAGUGACGAAUGUACGUCCUGGCCGGGCCAAGACCCCUCAUCCC-------------------- .....((((............(((((((...))))))).(((((..((((.((((......))))))))..)))))......)))).....-------------------- ( -35.60, z-score = -5.23, R) >droYak2.chr3R 4124174 90 - 28832112 UAUUAGAGGCACAUAAUAAACACAUCACCAAGUGAUGUCGGCCCA-CCAGUGACGAAUGCUCGCCCUGGCCGGGCCAAGACCCCUCCUCCC-------------------- .....((((............(((((((...))))))).(((((.-((((.(.(((....))).)))))..)))))......)))).....-------------------- ( -31.10, z-score = -3.65, R) >droEre2.scaffold_4770 11484723 90 + 17746568 UAUUAGAGGCACAUAAUAAACACAUCACCAAGUGAUGUCUCCCCA-CCACUGACGAAUGCUCGUCCUAGCCGGGCCAAGACCCCUCCUCCC-------------------- .......(((...........(((((((...))))))).......-.....(((((....)))))...)))(((......)))........-------------------- ( -20.70, z-score = -2.17, R) >droAna3.scaffold_13340 8673666 90 + 23697760 UAUUAGGGGCCCAUAAUAAACACAUCACCAAGUGAUGUCUGCUCG-GCAGUGACGAAUGUUCGGUAUGGCCAUGCCAGACCAGGCCGACCC-------------------- .....(((.............(((((((...)))))))....(((-((......((....))(((.((((...)))).)))..))))))))-------------------- ( -27.00, z-score = -0.68, R) >droWil1.scaffold_181130 4200972 91 - 16660200 UAUUAGAGGCACAUAAUAAACACAUCACCAAGUGAUGUCGACUUG---GGAGACGAAUGUUGACGAAUGUUGGCCCUGGCUAACCUUAUAUCCC----------------- .....((((.((((.........(((((...)))))((((((((.---(....).)).))))))..))))((((....)))).)))).......----------------- ( -22.80, z-score = -1.58, R) >droMoj3.scaffold_6540 21608600 111 + 34148556 UAUUAGAGGCCCAUAAUAAACACAUCACCAAGUGCUGUCGACUGAGGCAGUGACGAAUAUUUAACAAGACUGCCCAGACACGCCUCUGCAUGCCUCUGCGCCCCCCCCCCC ....((((((...........(((.(((...))).)))...(((.((((((.................)))))))))....))))))(((......)))............ ( -26.03, z-score = -2.29, R) >droGri2.scaffold_15116 715378 96 + 1808639 UAUUAGAGGCACAUAAUAAACACAUCACCAAGUGAUGUCGACUGAGGCAGUGACGAAUGUUUAACAAGGCUGGCCAGGCACGCAGCCCUCAACCCU--------------- .....(((........((((((((((((...))))))(((((((...))))..))).))))))....(((((..........))))))))......--------------- ( -21.90, z-score = -0.10, R) >droVir3.scaffold_13047 3005525 100 + 19223366 UAUUAGGGGCACAUAAUAAACACAUCAGCAAGUGGUGUCGACUGAGGCAGUGACGAAUGUUUAACAAGACUGGCCAGGCACGUC-CUCCGU-CCUUCACCCA--------- .....(((.............((((((.....)))))).(((.((((..(((......((((....))))........)))..)-))).))-).....))).--------- ( -22.64, z-score = 0.88, R) >consensus UAUUAGAGGCACAUAAUAAACACAUCACCAAGUGAUGUCGGCCCA_CCAGUGACGAAUGUUCGUCAUGGCCGGGCCAAGACCCCUCAUCCC____________________ .......(((............((((((...))))))(((.............)))............)))........................................ ( -7.08 = -7.27 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:58 2011