| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,360,312 – 15,360,426 |
| Length | 114 |
| Max. P | 0.890929 |

| Location | 15,360,312 – 15,360,426 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Shannon entropy | 0.33924 |
| G+C content | 0.52837 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15360312 114 + 27905053 AAAAUUUUUAUCAGCAUGCCGAGGCAAUGACAUGGCCACAUCACAAGGACGCGCAUGAGGGAGCGGGUUUUCGAGAUGGGA--GUGGGGAGCUUUGAGUGUGGAGGAUGUGGACAC ....((..((((..(((((((((((.....(((..((.((((.(.(((((.(((........))).))))).).)))))).--)))....)))))).)))))...))))..))... ( -33.60, z-score = -1.67, R) >droSim1.chr3R 21404163 114 - 27517382 AAAAUUUUUAUCAGCAUGCCGAGGCAAUGACAUGGCCACAUCACAAGGACGCGCACGAGGGAGCGGGUUUUUGUGAUGGGA--GGGGGGAGUGUGGAGUGCGGAGGAUGUGGACAC ..........((.((((.((...(((.(.((((..((.((((((((((((.(((........))).))))))))))))...--...))..)))).)..)))...)))))).))... ( -37.80, z-score = -3.29, R) >droSec1.super_25 411795 116 - 827797 AAAUUUUUUAUCAGCAUGCCGAGGCAAUGACAUGGCCACAUCACAAGGACGCGCACGAGGGAGCGGGUUUUCGAGAUGGGAGUGGGGGGAGUGAGGAGUGUGGAGAAUGAGGACAC ...(((((..((.((((.((...((..(..(((..((.((((.(.(((((.(((........))).))))).).)))))).)))..)...))..)).)))).))....)))))... ( -27.30, z-score = -0.35, R) >droYak2.chr3R_random 2405853 100 + 3277457 AAAAUUUUUAUCAGCAUGCCGAGGCAAUGACGUGGCCACAUCACACGGACGCGUAUGAGGGAGCGGGUUUUUCGGAUGGGAGUGUGGAGGAUGUGGACAC---------------- ..........(((...(((....))).))).(((.(((((((.((((..(.(((.(((((((.....))))))).))).)..))))...))))))).)))---------------- ( -29.90, z-score = -1.50, R) >droEre2.scaffold_4770 11465349 98 - 17746568 AAAAUUUUUAUCAGCAUGCCGAGGCAAUGACAUGACCACAUCACAAGGACGCGUAUGAGGGAGCGGGAUUUUGGGUUGGGA-UGUGG-GGAUAUGGACAC---------------- ..........(((...(((....))).)))((((.(((((((.(((((.(.(((........))).).)))))......))-)))))-...)))).....---------------- ( -23.50, z-score = -1.07, R) >consensus AAAAUUUUUAUCAGCAUGCCGAGGCAAUGACAUGGCCACAUCACAAGGACGCGCAUGAGGGAGCGGGUUUUUGAGAUGGGA_UGUGGGGAGUGUGGAGUG_GGAG_AUG_GGACAC ..........(((.(((((....)).)))...)))((.((((.(((((((.(((........))).))))))).)))))).................................... (-20.20 = -21.00 + 0.80)

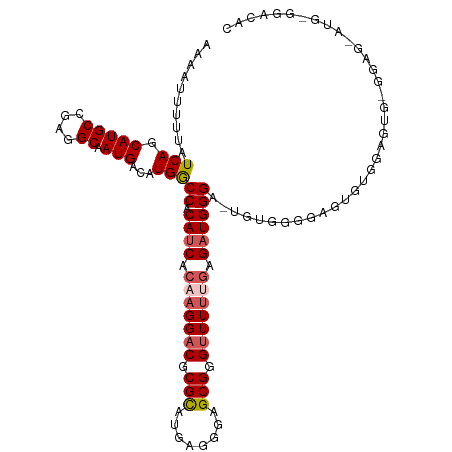

| Location | 15,360,312 – 15,360,426 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Shannon entropy | 0.33924 |
| G+C content | 0.52837 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -14.18 |
| Energy contribution | -15.34 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15360312 114 - 27905053 GUGUCCACAUCCUCCACACUCAAAGCUCCCCAC--UCCCAUCUCGAAAACCCGCUCCCUCAUGCGCGUCCUUGUGAUGUGGCCAUGUCAUUGCCUCGGCAUGCUGAUAAAAAUUUU ((((...........))))..............--.((((((.(((..((.(((........))).))..))).)))).))...(((((.(((....)))...)))))........ ( -21.70, z-score = -1.24, R) >droSim1.chr3R 21404163 114 + 27517382 GUGUCCACAUCCUCCGCACUCCACACUCCCCCC--UCCCAUCACAAAAACCCGCUCCCUCGUGCGCGUCCUUGUGAUGUGGCCAUGUCAUUGCCUCGGCAUGCUGAUAAAAAUUUU ((((...........))))..............--.((((((((((..((.(((........))).))..)))))))).))...(((((.(((....)))...)))))........ ( -26.30, z-score = -3.19, R) >droSec1.super_25 411795 116 + 827797 GUGUCCUCAUUCUCCACACUCCUCACUCCCCCCACUCCCAUCUCGAAAACCCGCUCCCUCGUGCGCGUCCUUGUGAUGUGGCCAUGUCAUUGCCUCGGCAUGCUGAUAAAAAAUUU ((((...........)))).................((((((.(((..((.(((........))).))..))).)))).))...(((((.(((....)))...)))))........ ( -21.70, z-score = -2.13, R) >droYak2.chr3R_random 2405853 100 - 3277457 ----------------GUGUCCACAUCCUCCACACUCCCAUCCGAAAAACCCGCUCCCUCAUACGCGUCCGUGUGAUGUGGCCACGUCAUUGCCUCGGCAUGCUGAUAAAAAUUUU ----------------(((.(((((((....((((((......))......(((..........)))...))))))))))).)))((((.(((....)))...))))......... ( -23.80, z-score = -2.50, R) >droEre2.scaffold_4770 11465349 98 + 17746568 ----------------GUGUCCAUAUCC-CCACA-UCCCAACCCAAAAUCCCGCUCCCUCAUACGCGUCCUUGUGAUGUGGUCAUGUCAUUGCCUCGGCAUGCUGAUAAAAAUUUU ----------------.......((((.-(((((-((.(((..........(((..........)))...))).))))))).((((((........))))))..))))........ ( -18.12, z-score = -1.41, R) >consensus GUGUCC_CAU_CUCC_CACUCCACACUCCCCACA_UCCCAUCCCAAAAACCCGCUCCCUCAUGCGCGUCCUUGUGAUGUGGCCAUGUCAUUGCCUCGGCAUGCUGAUAAAAAUUUU ....................................((((((.(((..((.(((........))).))..))).)))).))...(((((.(((....)))...)))))........ (-14.18 = -15.34 + 1.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:56 2011