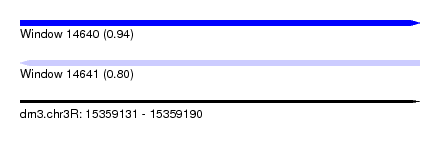
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,359,131 – 15,359,190 |
| Length | 59 |
| Max. P | 0.938194 |

| Location | 15,359,131 – 15,359,190 |
|---|---|
| Length | 59 |
| Sequences | 6 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 76.29 |
| Shannon entropy | 0.46447 |
| G+C content | 0.40051 |
| Mean single sequence MFE | -12.82 |
| Consensus MFE | -7.54 |
| Energy contribution | -8.60 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
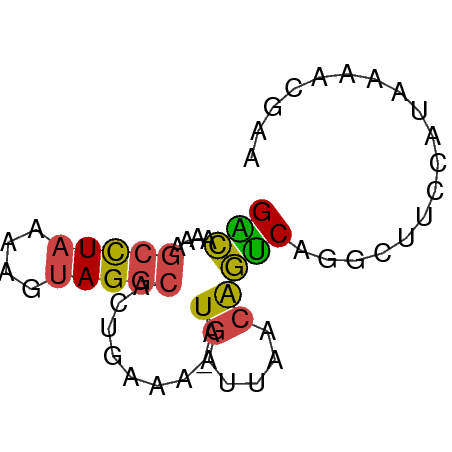
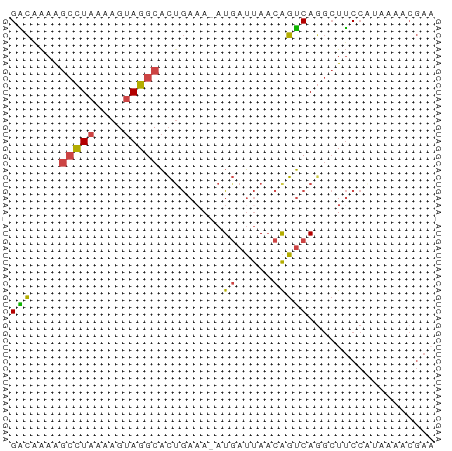
>dm3.chr3R 15359131 59 + 27905053 UUCGUUUUACGGAAGCUUGACUGUUAAUCAU-UUUCAGUGCCUACUUUUAGGCUUUGGUC ........((.(((((((((..((....(((-.....)))...))..))))))))).)). ( -12.80, z-score = -1.65, R) >droEre2.scaffold_4770 11464203 59 - 17746568 GUCGCUUUAUGGAAGCCUGACUGUUAAUCAU-UUUCAGUGCCUACUUUUAGGCUUUUGUC ........(..(((((((((..((....(((-.....)))...))..)))))))))..). ( -14.80, z-score = -2.41, R) >droYak2.chr3R 4118602 59 + 28832112 UUCGUUUUAUGGAAGCCUGACUGUUAAUCAU-UGUCCGUGCCUACUUUUAGGCUUUUGCC ..........((((((((((..((....(((-.....)))...))..))))))))))... ( -12.90, z-score = -1.84, R) >droSec1.super_25 410630 59 - 827797 UUCGUUUUAUGGAAGCCUGACUGUUAAUCAU-UUUCAGUGCCUACUUUUAGGCUUUUGUC ........(..(((((((((..((....(((-.....)))...))..)))))))))..). ( -14.80, z-score = -2.92, R) >droSim1.chr3R 21402984 59 - 27517382 UUCGUUUUAUGGAAGCCUGACUGUUAAUCAU-UUUCAGUGCCUACUUUUAGGCUUUUGCC ..........((((((((((..((....(((-.....)))...))..))))))))))... ( -13.40, z-score = -2.00, R) >droGri2.scaffold_15116 692868 50 - 1808639 ----------UGUAUUGUGUGCGGUGCUUAUGUUUUACAGCCUGCUUUUAACUCUCACAC ----------.....((((.((((.(((((.....)).)))))))..........)))). ( -8.20, z-score = 0.05, R) >consensus UUCGUUUUAUGGAAGCCUGACUGUUAAUCAU_UUUCAGUGCCUACUUUUAGGCUUUUGUC ..........((((((((((..((....(((......)))...))..))))))))))... ( -7.54 = -8.60 + 1.06)

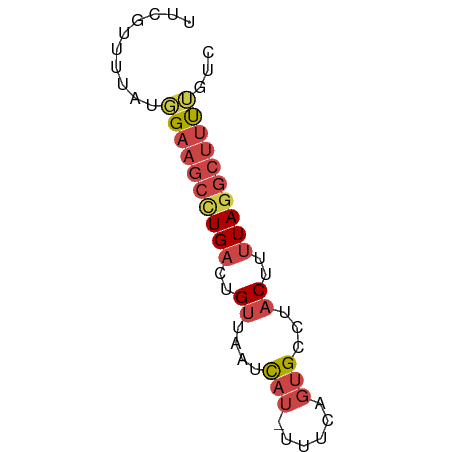
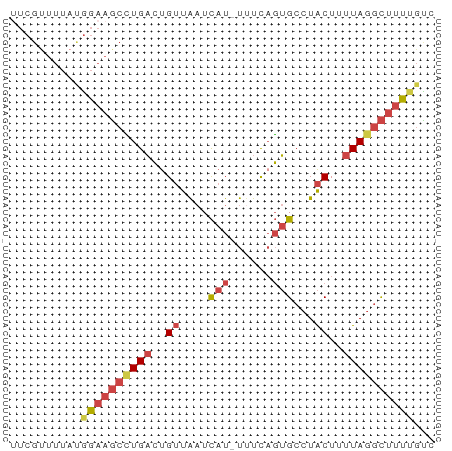
| Location | 15,359,131 – 15,359,190 |
|---|---|
| Length | 59 |
| Sequences | 6 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 76.29 |
| Shannon entropy | 0.46447 |
| G+C content | 0.40051 |
| Mean single sequence MFE | -11.30 |
| Consensus MFE | -5.63 |
| Energy contribution | -5.30 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
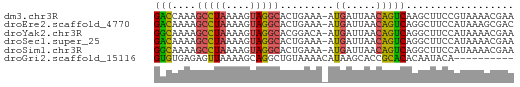
>dm3.chr3R 15359131 59 - 27905053 GACCAAAGCCUAAAAGUAGGCACUGAAA-AUGAUUAACAGUCAAGCUUCCGUAAAACGAA .......(((((....)))))((.(((.-.((((.....))))...))).))........ ( -9.50, z-score = -1.47, R) >droEre2.scaffold_4770 11464203 59 + 17746568 GACAAAAGCCUAAAAGUAGGCACUGAAA-AUGAUUAACAGUCAGGCUUCCAUAAAGCGAC .......(((((....))))).((((..-.((.....)).))))((((.....))))... ( -13.50, z-score = -2.75, R) >droYak2.chr3R 4118602 59 - 28832112 GGCAAAAGCCUAAAAGUAGGCACGGACA-AUGAUUAACAGUCAGGCUUCCAUAAAACGAA .......(((((....)))))..(((..-.((((.....))))....))).......... ( -12.30, z-score = -1.91, R) >droSec1.super_25 410630 59 + 827797 GACAAAAGCCUAAAAGUAGGCACUGAAA-AUGAUUAACAGUCAGGCUUCCAUAAAACGAA .......(((((....))))).((((..-.((.....)).))))................ ( -10.80, z-score = -1.99, R) >droSim1.chr3R 21402984 59 + 27517382 GGCAAAAGCCUAAAAGUAGGCACUGAAA-AUGAUUAACAGUCAGGCUUCCAUAAAACGAA (((....(((((....))))).((((..-.((.....)).)))))))............. ( -13.00, z-score = -2.17, R) >droGri2.scaffold_15116 692868 50 + 1808639 GUGUGAGAGUUAAAAGCAGGCUGUAAAACAUAAGCACCGCACACAAUACA---------- (((((.(.((.....))..(((..........)))..).)))))......---------- ( -8.70, z-score = -0.45, R) >consensus GACAAAAGCCUAAAAGUAGGCACUGAAA_AUGAUUAACAGUCAGGCUUCCAUAAAACGAA (((....(((((....))))).........((.....))))).................. ( -5.63 = -5.30 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:54 2011