
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,346,006 – 15,346,099 |
| Length | 93 |
| Max. P | 0.801520 |

| Location | 15,346,006 – 15,346,099 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.65 |
| Shannon entropy | 0.53247 |
| G+C content | 0.49334 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -18.43 |
| Energy contribution | -19.51 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
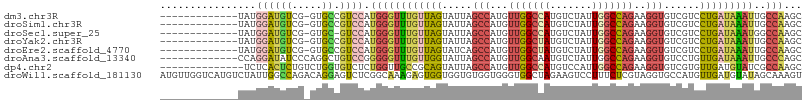
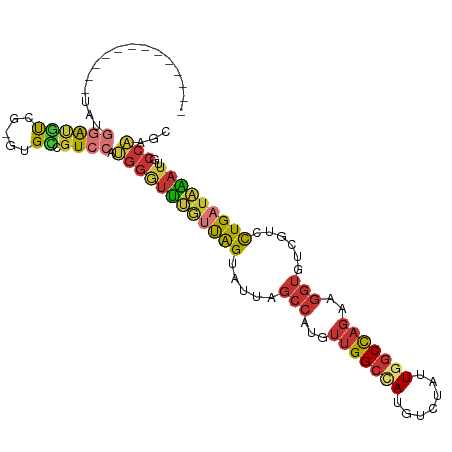
>dm3.chr3R 15346006 93 + 27905053 -------------UAUGGAUGUCG-GUGCCGUCCAUGGGUUUGUUAGUAUUAGCCAUGUUGGCCAUGUCUAUUGGCCAGAAGGUGUCGUCCUGAUAAAUUGCCAAGC -------------((((((((...-....))))))))((((((((((.((..(((...(((((((.......)))))))..)))...)).))))))))))....... ( -30.80, z-score = -2.04, R) >droSim1.chr3R 21389783 93 - 27517382 -------------UAUGGAUGUCG-GUGCCGUCCAUGGGUUUGUUAGUAUUAGCCAUGUUGGCCAUGUCUAUUGGCCAGAAGGUGUCGUCCUGAUAAAUUGCCAAGC -------------((((((((...-....))))))))((((((((((.((..(((...(((((((.......)))))))..)))...)).))))))))))....... ( -30.80, z-score = -2.04, R) >droSec1.super_25 397548 92 - 827797 -------------UAUGGAUGUCG-GUGC-GUCCAUGGGUUUGUUAGUAUUAGCCAUGUUGGCCAUGUCUAUUGGCCAGAAGGUGUCGUCCUGAUAAAUGGCCAAGC -------------(((((((((..-..))-)))))))((((((((((.((..(((...(((((((.......)))))))..)))...)).)))))))...))).... ( -32.50, z-score = -2.42, R) >droYak2.chr3R 4104842 93 + 28832112 -------------UAUGGAUGUCG-GUGCCGUCCAUGGGUUUGUUAGUAUUAGCCAUGUUGGCUAUGUCUAUUGGCCAGAAGGUGUCGUCCUGAUAAAUUGCCAAGC -------------((((((((...-....))))))))((((((((((.((..(((...(((((((.......)))))))..)))...)).))))))))))....... ( -28.50, z-score = -1.49, R) >droEre2.scaffold_4770 11450644 93 - 17746568 -------------UAUGGAUGUCG-GUGCCGUCCAUGGGUUUGUUAGUAUCAGCCAUGUUGGCUAUGUCUAUUGGCCAGAAGGUGUCGUCCUGAUAAAUUGCCAAGC -------------((((((((...-....))))))))((((((((((.((..(((...(((((((.......)))))))..)))...)).))))))))))....... ( -28.50, z-score = -1.30, R) >droAna3.scaffold_13340 8642864 94 - 23697760 -------------CCAGGAUAUCCCAGGCUGUCCGGGGGUUUGUUGGUAUUAGCCAUGUUGGCAAUGUCUAUUGGCCAGAAGGUGUCCUGUUGAUAAAUUGCCCAGC -------------.(((((((((((.((....)).))(((..((.(..(((.(((.....))))))..).))..)))....)))))))))................. ( -27.50, z-score = 0.32, R) >dp4.chr2 29830354 93 + 30794189 --------------UCUCACUCUGUCUGGUGUCUCUGGUUGCCGCAGUAUUAGCCAUGUUGGCCAUGUCCAUUGGCCAGAAGGUGUCGUGUUGAUGUAUCGCCAAGC --------------...((((......))))....(((((((..((((((..(((...(((((((.......)))))))..)))...))))))..)))..))))... ( -23.80, z-score = 0.64, R) >droWil1.scaffold_181130 4160902 107 + 16660200 AUGUUGGUCAUGUCUAUUGGCCAGACAGGAGUCUCGGCAAAGAGUGGUGGUGUGGUGGGUGGCUAGAAGUCCUUUCUCGUAGGUGCCAUGUUGAUGUAUAGCAAAGU ((((..(.(((((((((..(((((((....)))).)))...(((.((...(.((((.....)))).)...))...))))))))...)))))..)))).......... ( -25.00, z-score = 1.13, R) >consensus _____________UAUGGAUGUCG_GUGCCGUCCAUGGGUUUGUUAGUAUUAGCCAUGUUGGCCAUGUCUAUUGGCCAGAAGGUGUCGUCCUGAUAAAUUGCCAAGC ................(((((........))))).((((((((((((.....(((...(((((((.......)))))))..)))......)))))))))..)))... (-18.43 = -19.51 + 1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:52 2011