
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,342,802 – 15,342,902 |
| Length | 100 |
| Max. P | 0.600357 |

| Location | 15,342,802 – 15,342,902 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.66 |
| Shannon entropy | 0.30572 |
| G+C content | 0.51924 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.560582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
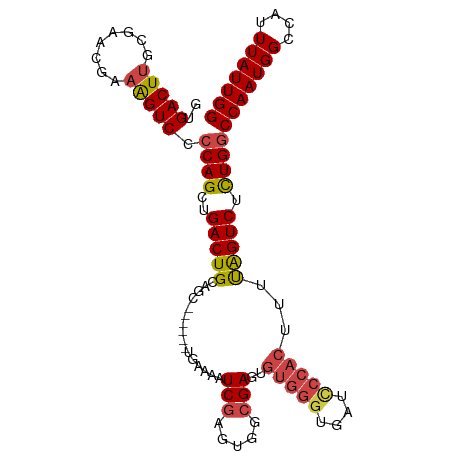
>dm3.chr3R 15342802 100 + 27905053 GUGACUUGCGAACGAAAGUCCACAGCUGACUGCAGC------UGAAUAUCGAGUGGCGAGUGUGGGUGAUUCCACUUUUAGUCUCUGGCCAAUGGCCAUUUAUUGG ((.(((((.(.((....)).).((((((....))))------)).....))))).))((.((..((((....))))..)).))..(((((...)))))........ ( -32.80, z-score = -1.59, R) >droSim1.chr3R 21386309 100 - 27517382 GUGACUUGCGAACGAAAGUCCCCAGCUGACUGCAGC------UGAAUAUCGAGUGGCGAGUGUGGGUGAUCCCACUUUUAGUCUCUGGCCAAUGGCCAUUUAUUGG ((.(((((.(.((....)).).((((((....))))------)).....))))).))(((.(((((....))))).......)))(((((...)))))........ ( -33.70, z-score = -1.81, R) >droSec1.super_25 394345 100 - 827797 GUGACUUGCGAACGAAAGUCCCCAGCUGACUGCAGC------UGAAUAUCGAGUGGCGAGUGUGGGUGAUCCCACUUUUAGUCUCUGGCCAAUGGCCAUUUAUUGG ((.(((((.(.((....)).).((((((....))))------)).....))))).))(((.(((((....))))).......)))(((((...)))))........ ( -33.70, z-score = -1.81, R) >droYak2.chr3R 4082396 106 + 28832112 GUGACUUGCGAGAGAAAGUCCCCAGCUGACUGCAAUGAGUGGCGAAAAUCGAGUGGCGAGUGUGGGUGAUCCCACUUUUAGUCUCUGGCCAAUGGCCAUUUAUUGG ..(((((.(....).)))))..(((....)))(((((((((((.(....(.((.(((....(((((....))))).....))).)).)....).))))))))))). ( -36.10, z-score = -1.81, R) >droEre2.scaffold_4770 11447602 100 - 17746568 GUGACUUGCGAGCGAAAGUCCCCAGCUGACUGCAGC------UGAAAAUCGAGUGUCGAGUGUGGGUGAUCCCACUUUUAGUCUCUGGCCAAUGGCCAUUUAUUGG ..(((...(((((....))...((((((....))))------))....)))...)))(((.(((((....))))).......)))(((((...)))))........ ( -33.30, z-score = -1.82, R) >droAna3.scaffold_13340 8639874 82 - 23697760 AUGACUUGCCGGCUGUGGUCCCCAGAUGACUGCAGC--------AGAAUCC--UUACGAGUGU--------------UCGGUCCUUGGCCAAUGGCCAUUUAUUGG .......(((.((((..(((.......)))..))))--------.(((..(--(....))..)--------------)))))...(((((...)))))........ ( -25.60, z-score = -0.82, R) >consensus GUGACUUGCGAACGAAAGUCCCCAGCUGACUGCAGC______UGAAAAUCGAGUGGCGAGUGUGGGUGAUCCCACUUUUAGUCUCUGGCCAAUGGCCAUUUAUUGG ..(((((........))))).((((..(((((................(((.....)))..(((((....)))))...))))).))))(((((((....))))))) (-21.99 = -22.28 + 0.30)


| Location | 15,342,802 – 15,342,902 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.66 |
| Shannon entropy | 0.30572 |
| G+C content | 0.51924 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -19.81 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
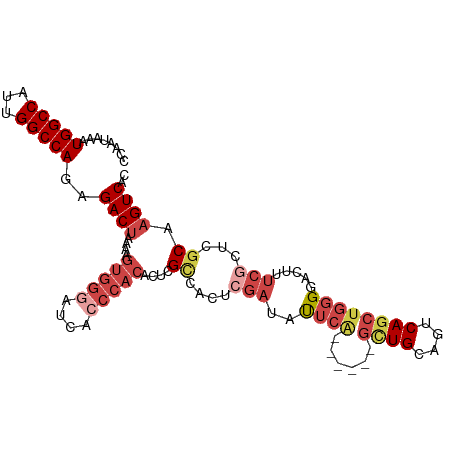
>dm3.chr3R 15342802 100 - 27905053 CCAAUAAAUGGCCAUUGGCCAGAGACUAAAAGUGGAAUCACCCACACUCGCCACUCGAUAUUCA------GCUGCAGUCAGCUGUGGACUUUCGUUCGCAAGUCAC ........(((((...)))))..((((....((((......))))....((...........((------((((....)))))).((((....)))))).)))).. ( -28.90, z-score = -2.01, R) >droSim1.chr3R 21386309 100 + 27517382 CCAAUAAAUGGCCAUUGGCCAGAGACUAAAAGUGGGAUCACCCACACUCGCCACUCGAUAUUCA------GCUGCAGUCAGCUGGGGACUUUCGUUCGCAAGUCAC ........(((((...)))))..((((....(((((....)))))....((....(((..((((------((((....)))))))).....)))...)).)))).. ( -33.40, z-score = -2.69, R) >droSec1.super_25 394345 100 + 827797 CCAAUAAAUGGCCAUUGGCCAGAGACUAAAAGUGGGAUCACCCACACUCGCCACUCGAUAUUCA------GCUGCAGUCAGCUGGGGACUUUCGUUCGCAAGUCAC ........(((((...)))))..((((....(((((....)))))....((....(((..((((------((((....)))))))).....)))...)).)))).. ( -33.40, z-score = -2.69, R) >droYak2.chr3R 4082396 106 - 28832112 CCAAUAAAUGGCCAUUGGCCAGAGACUAAAAGUGGGAUCACCCACACUCGCCACUCGAUUUUCGCCACUCAUUGCAGUCAGCUGGGGACUUUCUCUCGCAAGUCAC ........(((((...)))))..((((....(((((....)))))..((.(((((.((((...((........)))))))).))).))............)))).. ( -27.50, z-score = -0.74, R) >droEre2.scaffold_4770 11447602 100 + 17746568 CCAAUAAAUGGCCAUUGGCCAGAGACUAAAAGUGGGAUCACCCACACUCGACACUCGAUUUUCA------GCUGCAGUCAGCUGGGGACUUUCGCUCGCAAGUCAC ........(((((...)))))..((((....(((((....)))))...(((....(((((..((------((((....))))))..))...))).)))..)))).. ( -33.20, z-score = -2.67, R) >droAna3.scaffold_13340 8639874 82 + 23697760 CCAAUAAAUGGCCAUUGGCCAAGGACCGA--------------ACACUCGUAA--GGAUUCU--------GCUGCAGUCAUCUGGGGACCACAGCCGGCAAGUCAU .......(((((.....(((.((((((..--------------((....))..--)).))))--------((((..(((.......)))..)))).)))..))))) ( -20.20, z-score = 0.67, R) >consensus CCAAUAAAUGGCCAUUGGCCAGAGACUAAAAGUGGGAUCACCCACACUCGCCACUCGAUAUUCA______GCUGCAGUCAGCUGGGGACUUUCGCUCGCAAGUCAC ........(((((...)))))..((((....(((((....)))))..........................((.(((....))).)).............)))).. (-19.81 = -20.47 + 0.65)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:51 2011