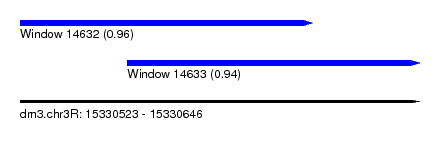
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,330,523 – 15,330,646 |
| Length | 123 |
| Max. P | 0.962903 |

| Location | 15,330,523 – 15,330,613 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.59 |
| Shannon entropy | 0.47227 |
| G+C content | 0.57696 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
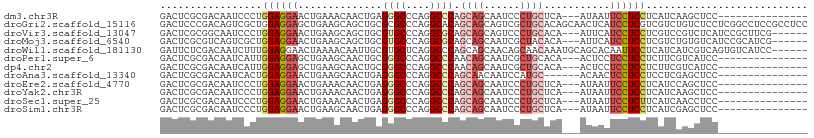
>dm3.chr3R 15330523 90 + 27905053 GACUCGCGACAAUCCCUGGAGGAACUGAAACAACUGAGGGCCCAGGCCCAGCAGCAAUCCCUGCUCA---AUAAUUCCUCCUCAUCAAGCUCC--------------- .....((..........(((((((.............((((....))))(((((......)))))..---....))))))).......))...--------------- ( -26.33, z-score = -1.95, R) >droGri2.scaffold_15116 653303 108 - 1808639 GACUCCCGACAGUCGCUGGAGGAGCUGAAGCAGCUGCGCGCCCAGGCACAGCAGCAGUCGCUGCACAGCAACUCAUCCUCGUCGUCUGUCUCCUCGGCCUCCGCCUCC .......(((((.((...(((((((....)).((((.(.((....)).).(((((....))))).))))......)))))..)).))))).....(((....)))... ( -39.90, z-score = -0.40, R) >droVir3.scaffold_13047 2951347 99 - 19223366 GACUCGCGGCAAUCCCUGGAGGAACUGAAGCAGCUGCGUGCCCAGGCGCAGCAGCAGUCCCUGCACA---AUUCAUCCUCCUCGUCCGUCUCAUCCGCUUCG------ .....((((........((((((..((((...(((((((......))))))).((((...))))...---.)))))))))).............))))....------ ( -33.10, z-score = -1.31, R) >droMoj3.scaffold_6540 21551944 99 - 34148556 GACUCGCGUCAGUCGCUGGAGGAACUGAAGCAGCUGCGUGCCCAGGCGCAGCAGCAAUCGCUACACA---AUUCAUCCUCCUCGUCUGUGUCAUCCGCAUCG------ .....(((.(((.((..((((((..((((...(((((((......)))))))(((....))).....---.)))))))))).)).))).......)))....------ ( -34.70, z-score = -1.48, R) >droWil1.scaffold_181130 4142166 102 + 16660200 GAUUCUCGACAAUCUUUGGAGGAACUAAAACAAUUGCGUGCUCAGGCCCAGCAGCAACAGCAACAAAUGCAGCACAAUUCCUCAUCAUCGUCAGUGUCAUCC------ (((.((.(((........((((((.........((((.((((.......))))))))..(((.....))).......))))))......))))).)))....------ ( -22.34, z-score = -0.88, R) >droPer1.super_6 5177945 90 + 6141320 GACUCGCGACAAUCAUUGGAGGAGCUGAAGCAACUGCGGGCCCAGGCCCAACAGCAAUCGCUGCACA---ACUCCUCCUCCUCUUCGUCAUCC--------------- .......(((.......(((((((.((..((....))((((....))))..((((....))))..))---.)))))))........)))....--------------- ( -32.26, z-score = -3.08, R) >dp4.chr2 29808409 90 + 30794189 GACUCGCGACAAUCAUUGGAGGAGCUGAAGCAACUGCGGGCCCAGGCCCAACAGCAAUCGCUGCACA---ACUCCUCCUCCUCUUCGUCAUCC--------------- .......(((.......(((((((.((..((....))((((....))))..((((....))))..))---.)))))))........)))....--------------- ( -32.26, z-score = -3.08, R) >droAna3.scaffold_13340 8628552 87 - 23697760 GACUCGCGACAAUCACUGGAGGAACUGAAGCAACUGAGGGCCCAGGCCCAGCAACAAUCCAUGC------ACAACUCCUCCUCCUCGAGCUCC--------------- ((((((.(.....)...((((((..((..........((((....)))).(((........)))------.))..))))))....)))).)).--------------- ( -25.50, z-score = -1.58, R) >droEre2.scaffold_4770 11435418 90 - 17746568 GACUCGCGACAAUCCCUGGAGGAACUGAAACAACUGAGGGCCCAGGCCCAGCAGCAAUCCCUGCUCA---AUAAUUCCUCCUCAUCCAGCUCC--------------- .....((..........(((((((.............((((....))))(((((......)))))..---....))))))).......))...--------------- ( -26.33, z-score = -1.87, R) >droYak2.chr3R 4070125 90 + 28832112 GACUCGCGACAAUCCCUGGAGGAACUGAAACAACUGAGGGCCCAGGCCCAGCAGCAAUCCCUGCUCA---AUAAUUCCUCCUCAUCAAGCUCC--------------- .....((..........(((((((.............((((....))))(((((......)))))..---....))))))).......))...--------------- ( -26.33, z-score = -1.95, R) >droSec1.super_25 382071 90 - 827797 GACUCGCGACAAUCCCUGGAGGAACUGAAACAACUGAGGGCCCAGGCCCAGCAGCAAUCCCUGCUCA---AUAAUUCCUCUUCAUCAACCUCC--------------- .................(((((...((((........((((....))))(((((......)))))..---..........))))....)))))--------------- ( -23.00, z-score = -1.42, R) >droSim1.chr3R 21372533 90 - 27517382 GACUCGCGACAAUCCCUGGAGGAACUGAAGCAACUGAGGGCCCAGGCCCAGCAGCAAUCCCUGCUCA---AUAAUUCCUCCUCAUCGAGCUCC--------------- ((((((.((........(((((((.............((((....))))(((((......)))))..---....)))))))))..)))).)).--------------- ( -27.70, z-score = -1.40, R) >consensus GACUCGCGACAAUCCCUGGAGGAACUGAAGCAACUGAGGGCCCAGGCCCAGCAGCAAUCCCUGCACA___AUAAUUCCUCCUCAUCAAGCUCC_______________ .................((((((..............((((....)))).((((......))))...........))))))........................... (-19.21 = -19.76 + 0.55)

| Location | 15,330,556 – 15,330,646 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.96 |
| Shannon entropy | 0.47672 |
| G+C content | 0.60190 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.87 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
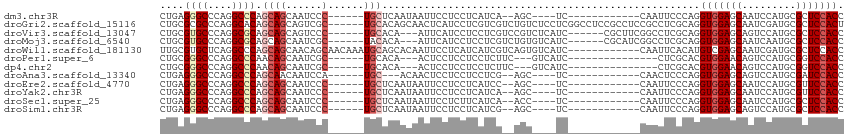
>dm3.chr3R 15330556 90 + 27905053 CUGAGGGCCCAGGCCCAGCAGCAAUCCC------UGCUCAAUAAUUCCUCCUCAUCA--AGC----UC------------CAAUUCCCAGGUGGAGCAAUCCAUGCGCUCCACC .(((((((....))))(((((......)------))))................)))--...----..------------.........((((((((.........)))))))) ( -29.10, z-score = -2.63, R) >droGri2.scaffold_15116 653336 108 - 1808639 CUGCGCGCCCAGGCACAGCAGCAGUCGC------UGCACAGCAACUCAUCCUCGUCGUCUGUCUCCUCGGCCUCCGCCUCCGCCUCGCAGGUGGAGCAAUCGAUGCGCUCCACU ..(((((((.(((....(((((....))------)))(((((.((........)).).))))..))).)))((((((((.((...)).))))))))........))))...... ( -40.50, z-score = -1.71, R) >droVir3.scaffold_13047 2951380 99 - 19223366 CUGCGUGCCCAGGCGCAGCAGCAGUCCC------UGCACA---AUUCAUCCUCCUCGUCCGUCUCAUC------CGCUUCGGCCUCGCAGGUGGAGCAGUCCAUGCGCUCCACC (((((.(((.(((((..((((......)------)))...---.............(.......)...------))))).)))..)))))(((((((.((....))))))))). ( -34.50, z-score = -2.11, R) >droMoj3.scaffold_6540 21551977 99 - 34148556 CUGCGUGCCCAGGCGCAGCAGCAAUCGC------UACACA---AUUCAUCCUCCUCGUCUGUGUCAUC------CGCAUCGGCCUCGCAGGUGGAGCAAUCAAUGCGCUCCACC (((((.(((...(((....(((....))------)(((((---................)))))....------)))...)))..)))))(((((((.........))))))). ( -32.99, z-score = -1.81, R) >droWil1.scaffold_181130 4142199 102 + 16660200 UUGCGUGCUCAGGCCCAGCAGCAACAGCAACAAAUGCAGCACAAUUCCUCAUCAUCGUCAGUGUCAUC------------CAAUUCACAUGUCGAGCAAUCGAUGCGCUCCACC (((((((((...((......))....(((.....))))))))............(((.(((((.....------------.....))).)).)))))))............... ( -19.50, z-score = 0.47, R) >droPer1.super_6 5177978 87 + 6141320 CUGCGGGCCCAGGCCCAACAGCAAUCGC------UGCACA---ACUCCUCCUCCUCUUC---GUCAUC---------------CUCGCACGUGGAACAGUCCAUGCGGUCCACC .(((((((....))))..((((....))------))....---................---......---------------...))).(((((...((....))..))))). ( -23.90, z-score = -1.19, R) >dp4.chr2 29808442 87 + 30794189 CUGCGGGCCCAGGCCCAACAGCAAUCGC------UGCACA---ACUCCUCCUCCUCUUC---GUCAUC---------------CUCGCACGUGGAACAGUCCAUGCGGUCCACC .(((((((....))))..((((....))------))....---................---......---------------...))).(((((...((....))..))))). ( -23.90, z-score = -1.19, R) >droAna3.scaffold_13340 8628585 87 - 23697760 CUGAGGGCCCAGGCCCAGCAACAAUCCA------UGC---ACAACUCCUCCUCCUCG--AGC----UC------------CAACUCCCAGGUGGAGCAGUCCAUGCGAUCCACC ....((((....)))).......(((((------((.---((..((((.(((....(--((.----..------------...)))..))).))))..)).)))).)))..... ( -27.40, z-score = -1.74, R) >droEre2.scaffold_4770 11435451 90 - 17746568 CUGAGGGCCCAGGCCCAGCAGCAAUCCC------UGCUCAAUAAUUCCUCCUCAUCC--AGC----UC------------CAAUUCCCAGGUGGAGCAAUCCAUGCGUUCCACC (((.((((....))))(((((......)------))))..................)--)).----..------------.........((((((((.........)))))))) ( -27.20, z-score = -2.29, R) >droYak2.chr3R 4070158 90 + 28832112 CUGAGGGCCCAGGCCCAGCAGCAAUCCC------UGCUCAAUAAUUCCUCCUCAUCA--AGC----UC------------CAAUUCCCAGGUGGAGCAAUCCAUGCGUUCCACC .(((((((....))))(((((......)------))))................)))--...----..------------.........((((((((.........)))))))) ( -26.70, z-score = -2.15, R) >droSec1.super_25 382104 90 - 827797 CUGAGGGCCCAGGCCCAGCAGCAAUCCC------UGCUCAAUAAUUCCUCUUCAUCA--ACC----UC------------CAAUUCCCAGGUGGAGCAAUCCAUGCGCUCCACC .(((((((....))))(((((......)------))))................)))--...----..------------.........((((((((.........)))))))) ( -29.10, z-score = -3.22, R) >droSim1.chr3R 21372566 90 - 27517382 CUGAGGGCCCAGGCCCAGCAGCAAUCCC------UGCUCAAUAAUUCCUCCUCAUCG--AGC----UC------------CAAUUCCCAGGUGGAGCAGUCCAUGCGCUCCACC .(((((((....)))).((((......)------)))))).......(((......)--)).----..------------.........((((((((.((....)))))))))) ( -29.40, z-score = -1.97, R) >consensus CUGAGGGCCCAGGCCCAGCAGCAAUCCC______UGCACAAUAAUUCCUCCUCAUCG__AGC____UC____________CAACUCCCAGGUGGAGCAAUCCAUGCGCUCCACC ....((((....))))..........................................................................(((((((.........))))))). (-16.68 = -16.87 + 0.18)
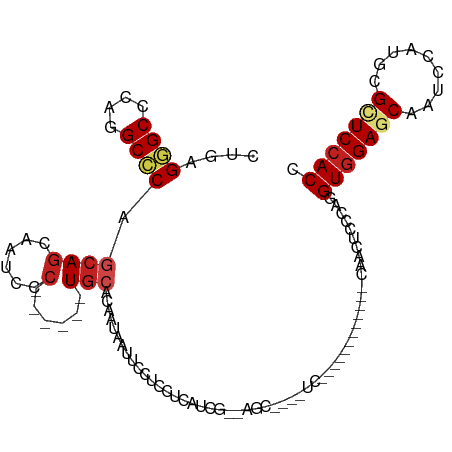
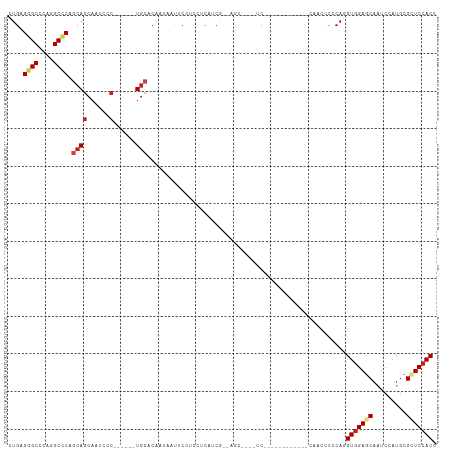
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:48 2011