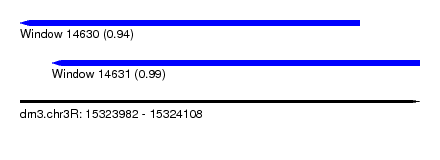
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,323,982 – 15,324,108 |
| Length | 126 |
| Max. P | 0.993428 |

| Location | 15,323,982 – 15,324,089 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.40 |
| Shannon entropy | 0.61484 |
| G+C content | 0.34974 |
| Mean single sequence MFE | -20.83 |
| Consensus MFE | -7.88 |
| Energy contribution | -7.99 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15323982 107 - 27905053 GCAAGUGCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAACGGCAUUUCGGUCAUAUGAGAUGCCCAGGGGCAAAAACAAAGACAGAG .....(((..(((.((((((((((((.....)))))))))))).................(((((((((......))))))))).))).)))............... ( -25.80, z-score = -3.25, R) >droSim1.chr3R 21365933 106 + 27517382 GCAAGUGCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAACGGCAUUUCGGUCAUAUGAGAUGGCCAGAGGCAAAAACAGAGACAGA- ((((((((........((((((((((.....))))))))))(((((......)))))....)))))).((((((.....))))))....))...............- ( -23.80, z-score = -2.66, R) >droYak2.chr3R 4060282 101 - 28832112 ----GUGUAAGC-UUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAGUGACAUUUCGGUCAUAUGAGAUGCCCAGAGAAAAAAACAGAAACAGA- ----((.....(-(((((((((((((.....))))))))))(((((......))))).(.(.(((((((......))))))).))))))......)).........- ( -15.30, z-score = -0.74, R) >droEre2.scaffold_4770 11429064 105 + 17746568 GCGAGUGCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAACGACAUUUCGGUCAUAUGAGAUGCCCAGAGG-ACAAACAGAGACAGA- ((....))..(((((.((((((((((.....))))))))))(((((......))))).....(((((((......)))))))...)))))-...............- ( -18.40, z-score = -1.39, R) >droAna3.scaffold_13340 8622267 99 + 23697760 ACGAGUGCAAGCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAGC-AAAAUUGUUGACAACACUU-GGUCAUACGAGAUGGCCAGACAGAAAAACAGAG------ ..(((((...(((.((((((((((((.....)))))))))))).)))-....(((....))))))))-((((((.....))))))................------ ( -25.40, z-score = -4.27, R) >droVir3.scaffold_13047 2943272 89 + 19223366 ------GCUAACGUUUUAAUGGAUUUAAUUAAAAUUCAUUUUAUCCUGGCCAUAGUGCCCAGCACAU-----GAACGAGGUACCAGG----AAUACCAGGAUUC--- ------...........(((((((((.....)))))))))..(((((((((...(((((..(.....-----...)..)))))..))----....)))))))..--- ( -20.30, z-score = -1.64, R) >droGri2.scaffold_15116 643845 95 + 1808639 --AGGCUCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUU-CACGAC-AGAAUUGUGCAUAGUGAAGAGAACAAUGGGGACAACUUGA---AUCGAUGGGGA----- --.(..((((.(((((((((((((((.....)))))))).-(((((.-....))))).....))))))).....((....))..))))---..)........----- ( -16.80, z-score = -0.28, R) >consensus _CAAGUGCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAACGGCAUUUCGGUCAUAUGAGAUGCCCAGAGG_AAAAACAGAGACAGA_ ....((((........((((((((((.....))))))))))(((((......)))))....)))).......................................... ( -7.88 = -7.99 + 0.11)

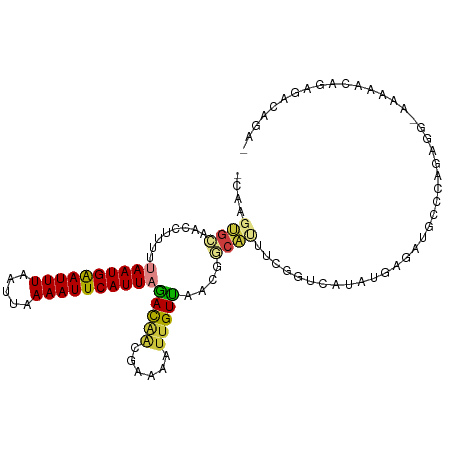
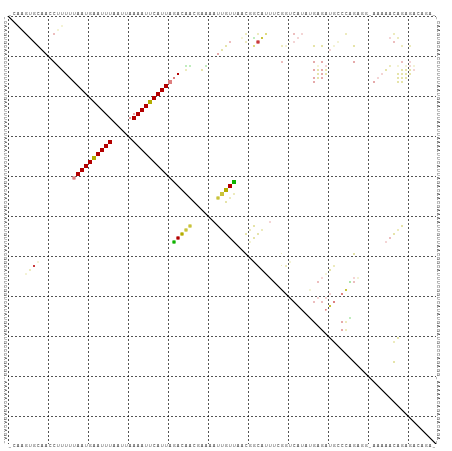
| Location | 15,323,992 – 15,324,108 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Shannon entropy | 0.30337 |
| G+C content | 0.36237 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.59 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15323992 116 - 27905053 UUAUGUCCUUGGGCACCCAGCAAGUGCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAACGGCAUUUCGGUCAUAUGAGAUGCCCAGGGGCAAAAAC ...((((((((((((.(((((....)).(((..((((((((((((.....))))))))))))....((((..(((.....))))))))))....)).).))))))))))))..... ( -37.80, z-score = -4.63, R) >droSim1.chr3R 21365942 116 + 27517382 UUAUGUCCUUGGGCACCCAGCAAGUGCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAACGGCAUUUCGGUCAUAUGAGAUGGCCAGAGGCAAAAAC ...((.((((.(((..((.((....))......((((((((((((.....)))))))))))).................))(((((((......)))))))))).))))))..... ( -29.10, z-score = -2.36, R) >droYak2.chr3R 4060291 111 - 28832112 UUUUACCCUUUGGGACGCAGUGUAAGC-----UUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAGUGACAUUUCGGUCAUAUGAGAUGCCCAGAGAAAAAAAC .......(((((((.(((.........-----...((((((((((.....))))))))))(((((......))))).))).(((((((......))))))))))))))........ ( -27.60, z-score = -3.51, R) >droEre2.scaffold_4770 11429073 115 + 17746568 UUAUGCCCUUUGGAGCCCAGCGAGUGCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAACGACAUUUCGGUCAUAUGAGAUGCCCAGAGGACAAAC- ...((.(((((((.((....((.(((..(((..((((((((((((.....))))))))))))....((((..((((...)))))))))))))).))....))))))))).))...- ( -29.40, z-score = -3.78, R) >droAna3.scaffold_13340 8622271 110 + 23697760 ---UGUCCUGCAGGAC-UAACGAGUGCAAGCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAGC-AAAAUUGUUGACAACACUU-GGUCAUACGAGAUGGCCAGACAGAAAAAC ---.((((....))))-....(((((...(((.((((((((((((.....)))))))))))).)))-....(((....))))))))-((((((.....))))))............ ( -30.60, z-score = -4.04, R) >consensus UUAUGUCCUUGGGCACCCAGCGAGUGCAACCUUUUUAAUGAAUUUAAUUAAAAUUCAUUAGACAACGAAAAUUGUUAACGACAUUUCGGUCAUAUGAGAUGCCCAGAGGAAAAAAC ......((((.((.....(((((..........((((((((((((.....)))))))))))).........))))).....(((((((......))))))).)).))))....... (-15.87 = -16.59 + 0.72)

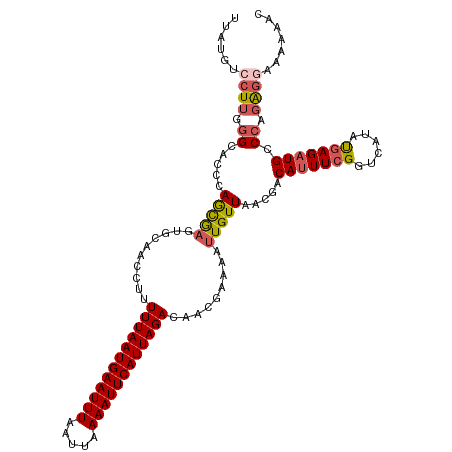
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:46 2011