| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,323,793 – 15,323,889 |
| Length | 96 |
| Max. P | 0.907335 |

| Location | 15,323,793 – 15,323,889 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.94 |
| Shannon entropy | 0.60167 |
| G+C content | 0.45032 |
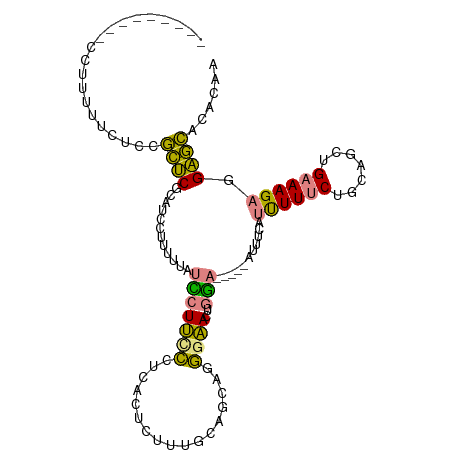
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -9.12 |
| Energy contribution | -7.66 |
| Covariance contribution | -1.46 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
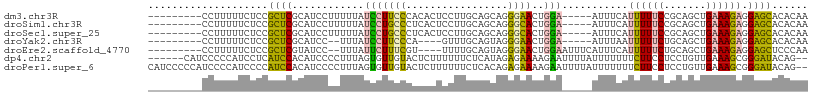
>dm3.chr3R 15323793 96 - 27905053 ---------CCUUUUUCUCCGCUCGCAUCCUUUUUAUCCUUCCCACACUCCUUGCAGCAGGGAACUGGA-----AUUUCAUUUUUCCGCAGCUGAAAGAGGAGCACACAA ---------((((((((...(((.((......................((((((...))))))...(((-----(........))))))))).))))))))......... ( -24.20, z-score = -2.13, R) >droSim1.chr3R 21365744 96 + 27517382 ---------CCUUUUUCUCCGCUCGCAUCCUUUUUAUCCUGCCCUCACUCCUUGCAGCAGGGCACUGGA-----AUUUCAUUUUUCCGCAGCUGAAAGAGGAGCACACAA ---------((((((((...(((.((.............((((((..((......)).))))))..(((-----(........))))))))).))))))))......... ( -27.10, z-score = -2.34, R) >droSec1.super_25 375342 96 + 827797 ---------CCUUUUUCUCCGCUCGCAUCCUUUUUAUCCUGCCCUCACUCCUUGCAGCAGGGCACUGGA-----AUUUCAUUUUUCCGCAGCUGAAAGAGGAGCACACAA ---------((((((((...(((.((.............((((((..((......)).))))))..(((-----(........))))))))).))))))))......... ( -27.10, z-score = -2.34, R) >droYak2.chr3R 4060091 90 - 28832112 ---------CCUUUUUCUCCGCUCGCAUCC--UUUAUCCUUCCCA----GUUUGCAGUAGGGAACUGGA-----AUUUAAUUUUUCUGCAGCUGAAAGAGGAGCACACAA ---------((((((((...(((.(((...--..........(((----((((.(....).))))))).-----............)))))).))))))))......... ( -24.47, z-score = -1.79, R) >droEre2.scaffold_4770 11428880 95 + 17746568 ---------CCUUUUUCUCCGCUCGUAUCC--UUUAUUCUUUCGU----UUUUGCAGUAGGGAACUGGAAUUUCAUUUCAUUUUUCUGCAGCUGAAAGAGGAGCUCCCAA ---------((((((((...(((.......--...........((----....)).((((..((.((((((...)))))).))..))))))).))))))))......... ( -22.00, z-score = -1.12, R) >dp4.chr2 29798548 102 - 30794189 ------CAUCCCCCAUCCUCAUCCACAUCCCCUUUAGUGUUGUACUCUUUUUUCUCAUAGAGAAAAGAAUUUUAUUUUUUUCUUCCUCCUGUUGAAAGCGGGAUACAG-- ------.................(((..........))).((((.(((((((((.....)))))))))..................((((((.....)))))))))).-- ( -16.90, z-score = -1.49, R) >droPer1.super_6 5168472 108 - 6141320 CAUCCCCCAUCCCCAUCCCCAUCCACAUCCCCUUUAGUGUUGUACUCUUUUUUCUCACAGAGAAAAGAAUUUUAUUUUUUUCUUCCUCCUGUUGAAAGCGGGAUACAG-- .......................(((..........))).((((.(((((((((.....)))))))))..................((((((.....)))))))))).-- ( -16.90, z-score = -1.73, R) >consensus _________CCUUUUUCUCCGCUCGCAUCCUUUUUAUCCUUCCCUCACUCUUUGCAGCAGGGAACUGGA_____AUUUCAUUUUUCUGCAGCUGAAAGAGGAGCACACAA ....................((((...............((((.................))))................((((((.......)))))).))))...... ( -9.12 = -7.66 + -1.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:45 2011