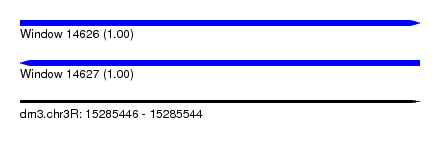
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,285,446 – 15,285,544 |
| Length | 98 |
| Max. P | 0.999620 |

| Location | 15,285,446 – 15,285,544 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.46 |
| Shannon entropy | 0.56078 |
| G+C content | 0.30976 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -18.14 |
| Energy contribution | -16.82 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.36 |
| Mean z-score | -4.24 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
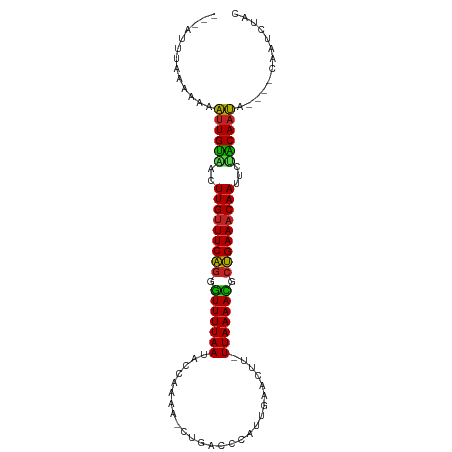
>dm3.chr3R 15285446 98 + 27905053 -UCAUUUAAACAAAUUGUUACUUGUUUCAGGGUUUUAAUACCAAAA-CUGACCCAUUGAACCU-UUAAAACGCUGAAACAAUUCAACAAUA--AGAAAUGUAC -.(((((......((((((..(((((((((.(((((((........-................-))))))).)))))))))...)))))).--..)))))... ( -21.06, z-score = -3.47, R) >droVir3.scaffold_13047 2896520 88 - 19223366 --------UCGUCUUUGUAACUUGAUUCAGUGUUUUAAGCCCAAAA-CUGACCCAAAA--CAAAUUAAAAUGCUGAAGCAAUUCUACAAAG----CAGUCCGC --------..(.(((((((..(((.(((((((((((((........-...........--....))))))))))))).)))...)))))))----)....... ( -19.00, z-score = -3.02, R) >droGri2.scaffold_15116 591679 91 - 1808639 --------UCGCAUUUGUGACUUGUUUCAGUAUUUUAAGCCCAAAAACUGACCCAAAAAACAAAUUAAAAUGCUGAAACAAUUUCGCAAAU----CUAUACAC --------..(.((((((((.(((((((((((((((((..........................)))))))))))))))))..))))))))----)....... ( -24.37, z-score = -6.33, R) >droMoj3.scaffold_6540 21495632 93 - 34148556 ---ACUUUGAAGCUCUGCAACUUGUUUCAGCAUUUUAAGCCCAAAA-CUGACCCAAUA--UUUAUUAAAAUGCUGAAACAAUUCUGCAAAA----CUUUUCAC ---....(((((...((((..(((((((((((((((((........-...........--....)))))))))))))))))...))))...----..))))). ( -24.20, z-score = -5.29, R) >droWil1.scaffold_181130 4085936 78 + 16660200 -------------UUUGUAACUUGUUUCAGUGUUUUAAAAACAAAA-CUGACCCAUUGAACUAUUUAAAAUGCUGAAACAAUUCUACAAAA----C------- -------------((((((..((((((((((((((((((.......-................))))))))))))))))))...)))))).----.------- ( -19.90, z-score = -5.30, R) >droPer1.super_6 5120768 87 + 6141320 -------AAAACAUUUGUAACUUGUUUCAGGGUUUUAA-ACCAAAA-CUGACCCAUUGAACUC-UUAAAAUCCUGAAACAAUUCUACAAAA------AUUUAC -------......((((((..(((((((((((((((((-.......-................-)))))))))))))))))...)))))).------...... ( -22.10, z-score = -5.27, R) >droAna3.scaffold_13340 8591146 97 - 23697760 ----UUUUAAUAAAUUGCAACUUGUUUCGGGGUUUUAAUACCAAAA-CUGACCCAUUGAACCU-UUAAAACGGCGAAACAAUUCUGCAAUAUUGACAGUCCAU ----..((((((..(((((..(((((((((((((............-..)))))......((.-.......))))))))))...)))))))))))........ ( -21.24, z-score = -2.54, R) >droYak2.chr3R 4017030 99 + 28832112 UCCAUUUUAAAAAAUUGUCACUUGUUUCAGGGUUUUAAUACCAAAA-CUGACCCAUUGAACAU-UUAAAACGCUGAAACAAUUUUACAAUA--GACAAUCUUC .............(((((...(((((((((.(((((((........-................-))))))).)))))))))....))))).--.......... ( -18.76, z-score = -2.99, R) >droEre2.scaffold_4770 11390583 99 - 17746568 UCCAUUCAACAAAAUUGUUACUUGUUUCAGGGUUUUAAUACCAAAA-CUGACCCAUUGAACUU-UUAAAACGCUGAAACAAUUUCACAAUA--GCCAAUCUUC .............(((((...(((((((((.(((((((........-................-))))))).)))))))))....))))).--.......... ( -18.76, z-score = -3.12, R) >droSec1.super_25 336695 98 - 827797 -UCAUUUAAACAAAUUGUUACUUGUUUCAGUGUUUUAAUGCCAAAA-CUGACCCAUUGAACUU-UUAAAACGCUGAAACAAUUCAACAAUA--AGAAAUCUAC -............((((((..(((((((((((((((((........-................-)))))))))))))))))...)))))).--.......... ( -22.26, z-score = -4.65, R) >droSim1.chr3R 21326039 98 - 27517382 -UCAUUUAAACAAAUUGUUACUUGUUUCAGUGUUUUAAUGCCAAAA-CUGACCCAUUGAACUU-UUAAAACGCUGAAACAAUUCAACAAUA--AGAAAUCUAC -............((((((..(((((((((((((((((........-................-)))))))))))))))))...)))))).--.......... ( -22.26, z-score = -4.65, R) >consensus ___AUUUAAAAAAAUUGUAACUUGUUUCAGGGUUUUAAUACCAAAA_CUGACCCAUUGAACUU_UUAAAACGCUGAAACAAUUCUACAAUA____CAAUCUAC .............((((((..(((((((((.(((((((..........................))))))).)))))))))...))))))............. (-18.14 = -16.82 + -1.33)

| Location | 15,285,446 – 15,285,544 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.46 |
| Shannon entropy | 0.56078 |
| G+C content | 0.30976 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -17.22 |
| Energy contribution | -16.16 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
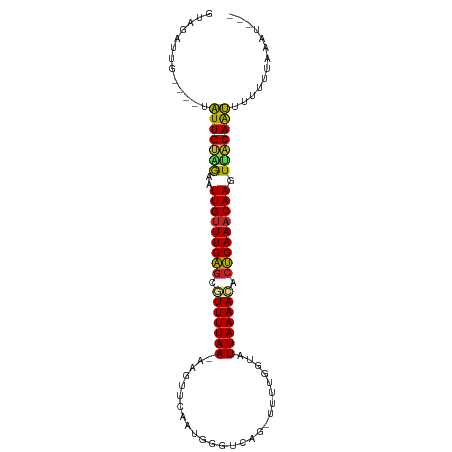
>dm3.chr3R 15285446 98 - 27905053 GUACAUUUCU--UAUUGUUGAAUUGUUUCAGCGUUUUAA-AGGUUCAAUGGGUCAG-UUUUGGUAUUAAAACCCUGAAACAAGUAACAAUUUGUUUAAAUGA- ...((((((.--.(((((((..(((((((((.(((((((-....((((........-..))))..))))))).))))))))).)))))))..)...))))).- ( -23.20, z-score = -2.48, R) >droVir3.scaffold_13047 2896520 88 + 19223366 GCGGACUG----CUUUGUAGAAUUGCUUCAGCAUUUUAAUUUG--UUUUGGGUCAG-UUUUGGGCUUAAAACACUGAAUCAAGUUACAAAGACGA-------- .((.....----((((((((..(((.(((((..........((--(((((((((..-.....)))))))))))))))).))).)))))))).)).-------- ( -22.90, z-score = -1.94, R) >droGri2.scaffold_15116 591679 91 + 1808639 GUGUAUAG----AUUUGCGAAAUUGUUUCAGCAUUUUAAUUUGUUUUUUGGGUCAGUUUUUGGGCUUAAAAUACUGAAACAAGUCACAAAUGCGA-------- ........----(((((.((..(((((((((((........))..(((((((((........)))))))))..))))))))).)).)))))....-------- ( -19.90, z-score = -1.40, R) >droMoj3.scaffold_6540 21495632 93 + 34148556 GUGAAAAG----UUUUGCAGAAUUGUUUCAGCAUUUUAAUAAA--UAUUGGGUCAG-UUUUGGGCUUAAAAUGCUGAAACAAGUUGCAGAGCUUCAAAGU--- .(((..((----((((((((..((((((((((((((((.....--.....((((..-.....)))))))))))))))))))).)))))))))))))....--- ( -33.20, z-score = -5.36, R) >droWil1.scaffold_181130 4085936 78 - 16660200 -------G----UUUUGUAGAAUUGUUUCAGCAUUUUAAAUAGUUCAAUGGGUCAG-UUUUGUUUUUAAAACACUGAAACAAGUUACAAA------------- -------.----.(((((((..(((((((((..(((((((.....(((........-..)))..)))))))..))))))))).)))))))------------- ( -16.90, z-score = -2.34, R) >droPer1.super_6 5120768 87 - 6141320 GUAAAU------UUUUGUAGAAUUGUUUCAGGAUUUUAA-GAGUUCAAUGGGUCAG-UUUUGGU-UUAAAACCCUGAAACAAGUUACAAAUGUUUU------- ..((((------.(((((((..((((((((((.((((((-(...((((........-..)))))-)))))).)))))))))).))))))).)))).------- ( -22.30, z-score = -3.13, R) >droAna3.scaffold_13340 8591146 97 + 23697760 AUGGACUGUCAAUAUUGCAGAAUUGUUUCGCCGUUUUAA-AGGUUCAAUGGGUCAG-UUUUGGUAUUAAAACCCCGAAACAAGUUGCAAUUUAUUAAAA---- .............(((((((..((((((((((.......-.))).....(((...(-(((((....)))))))))))))))).))))))).........---- ( -21.30, z-score = -1.24, R) >droYak2.chr3R 4017030 99 - 28832112 GAAGAUUGUC--UAUUGUAAAAUUGUUUCAGCGUUUUAA-AUGUUCAAUGGGUCAG-UUUUGGUAUUAAAACCCUGAAACAAGUGACAAUUUUUUAAAAUGGA ((((((((((--..........(((((((((.(((((((-....((((........-..))))..))))))).)))))))))..))))))))))......... ( -23.40, z-score = -2.50, R) >droEre2.scaffold_4770 11390583 99 + 17746568 GAAGAUUGGC--UAUUGUGAAAUUGUUUCAGCGUUUUAA-AAGUUCAAUGGGUCAG-UUUUGGUAUUAAAACCCUGAAACAAGUAACAAUUUUGUUGAAUGGA .....(..((--.(((((....(((((((((.(((((((-....((((........-..))))..))))))).)))))))))...)))))...))..)..... ( -24.00, z-score = -2.17, R) >droSec1.super_25 336695 98 + 827797 GUAGAUUUCU--UAUUGUUGAAUUGUUUCAGCGUUUUAA-AAGUUCAAUGGGUCAG-UUUUGGCAUUAAAACACUGAAACAAGUAACAAUUUGUUUAAAUGA- .(((((....--.(((((((..(((((((((.(((((((-.....(....)(((..-....))).))))))).))))))))).)))))))..))))).....- ( -22.80, z-score = -2.39, R) >droSim1.chr3R 21326039 98 + 27517382 GUAGAUUUCU--UAUUGUUGAAUUGUUUCAGCGUUUUAA-AAGUUCAAUGGGUCAG-UUUUGGCAUUAAAACACUGAAACAAGUAACAAUUUGUUUAAAUGA- .(((((....--.(((((((..(((((((((.(((((((-.....(....)(((..-....))).))))))).))))))))).)))))))..))))).....- ( -22.80, z-score = -2.39, R) >consensus GUAGAUUG____UAUUGUAGAAUUGUUUCAGCGUUUUAA_AAGUUCAAUGGGUCAG_UUUUGGUAUUAAAACACUGAAACAAGUUACAAUUUUUUUAAAU___ .............(((((((..(((((((((.(((((((..........................))))))).))))))))).)))))))............. (-17.22 = -16.16 + -1.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:43 2011