| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,189,217 – 15,189,317 |
| Length | 100 |
| Max. P | 0.996575 |

| Location | 15,189,217 – 15,189,317 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.46 |
| Shannon entropy | 0.55709 |
| G+C content | 0.51494 |
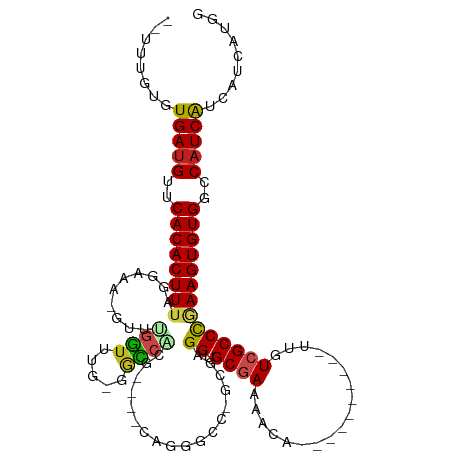
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -23.23 |
| Energy contribution | -22.76 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
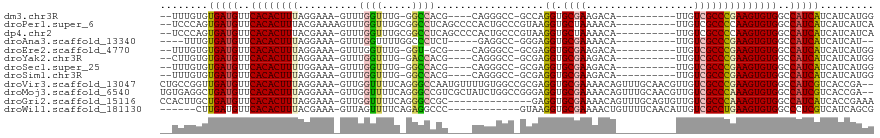
>dm3.chr3R 15189217 100 + 27905053 --UUUGUGUGAUGUUCACACUUUAGGAAA-GUUUGGUUUG-GGCCACG----CAGGGCC-GCCAGGUGCGAAGACA----------UUGUCGCCCGAAGUGUGGCCAUCAUCAUCAUGG --...((((((((..((((((((......-...((((...-((((...----...))))-))))((.((((.....----------...))))))))))))))..))))).)))..... ( -38.00, z-score = -2.10, R) >droPer1.super_6 5022110 107 + 6141320 --UCCCAGUGAUGUUCACACUUUACGAAAAGUUUGGUUUGCGGCCUCAGCCCCACUGCCCGUAAGGUGCUAAAACA----------UUGUCGCCCCAAGUGUGGCCAUCAUCAUCAUCA --.....((((((..(((((((..(((.((.((((((((((((...(((.....))).))))))...))))))...----------)).)))....)))))))..))))))........ ( -30.80, z-score = -2.18, R) >dp4.chr2 29651123 106 + 30794189 --UCCCAGUGAUGUUCACACUUUACGAAA-GUUUGGUUUGCGGCCUCAGCCCCACUGCCCGUAAGGUGCUAAAACA----------UUGUCGCCCCAAGUGUGGCCAUCAUCAUCAUCA --.....((((((..(((((...((....-))((((...(((((............((((....)).)).......----------..))))).)))))))))..))))))........ ( -32.10, z-score = -2.64, R) >droAna3.scaffold_13340 8496596 96 - 23697760 ----UUUGUGAUGUUCACACUUUAGGAAA-GUUUGGUUUUGGCCCUCU-----GAGGCC-GGGAGGUGCGAAAACA----------UUGUCGCCCGAAGUGUGGCCAUCAUCAUCAU-- ----...((((((..(((((((..((...-.....(((((.(((((((-----......-.))))).)).))))).----------......))..)))))))..))))))......-- ( -38.06, z-score = -3.39, R) >droEre2.scaffold_4770 11301127 99 - 17746568 --UUUGUGUGAUGUUCACACUUUAGGAAA-GUUUGGUUUG-GGU-GCG----CAGGGCC-GCGAGGUGCGAAGACA----------UUGUCGCCCGAAGUGUGGCCAUCAUCAUCAUGG --...((((((((..(((((((..((...-.....((((.-...-.((----(...(((-....)))))).)))).----------......))..)))))))..))))).)))..... ( -34.06, z-score = -1.24, R) >droYak2.chr3R 3911179 100 + 28832112 --CUUGUGUGAUGUUCACACUUUAGGAAA-GUUUGGUUUG-GACCACG----CAGGGCC-GCGAGGUGCGAAGACA----------UUGUCGCCCGAAGUGUGGCCAUCAUCAUCAUGG --...((((((((..((((((((......-((.((((...-.)))).)----).((((.-((((.((......)).----------)))).))))))))))))..))))).)))..... ( -37.30, z-score = -2.08, R) >droSec1.super_25 248625 100 - 827797 --UUUGUGUGAUGUUCACACUUUUGGAAA-GUUUGGUUUG-GGCCACG----CAGGGCC-GCGAGGUGCGAAGACA----------UUGUCGCCCGAAGUGUGGCCAUCAUCAUCAUGG --.(..(((((((.(((.((((.....))-)).)))....-(((((((----(.((((.-((((.((......)).----------)))).))))...))))))))..))))).))..) ( -37.40, z-score = -1.78, R) >droSim1.chr3R 21227945 100 - 27517382 --UUUGUGUGAUGUUCACACUUUAGGAAA-GUUUGGUUUG-GGCCACG----CAGGGCC-GCGAGGUGCGAAGACA----------UUGUCGCCCGAAGUGUGGCCAUCAUCAUCAUGG --...((((((((..((((((((......-((.((((...-.)))).)----).((((.-((((.((......)).----------)))).))))))))))))..))))).)))..... ( -37.70, z-score = -2.09, R) >droVir3.scaffold_13047 2779955 116 - 19223366 CUGCCGGUUGAUGUUCACACUUUAGGAAA-GUUGGUUUUCAGGGCCAAUGUUUUGUGGCCGCGAGGUGCGAAAACAGUUUGCAACGUUGUCGCCCGAAGUGUGGCCAUCGUCACCGA-- ....(((((((((..((((((((..((((-(....))))).(((((((((((.....(((....)))(((((.....))))))))))))..))))))))))))..)))))..)))).-- ( -46.30, z-score = -2.82, R) >droMoj3.scaffold_6540 21387936 116 - 34148556 UGUGAGGCUGAUGUUCACACUUUAGGAAA-GUUGGUUUUCAGGGCCGUCGCUAUCUGGCCGGGAGGUGCGAAAACAGUUUGCAACGUUGUCGCCCAAAGUGUGGCCAUCGUCACCGA-- .((((...(((((..((((((((......-........((..(((((........)))))..))((.((((.(((.((.....))))).))))))))))))))..)))))))))...-- ( -39.00, z-score = -0.48, R) >droGri2.scaffold_15116 429877 104 - 1808639 CCACUUGCUGAUGUUCACACUUUAGGAAA-GUUGGUUUUCAGGGCCGC--------------GAGGUGCGAAAACAGUUUGCAGUGUUGUCGCCCAAAGUGUGGCCAUCAUCACCGAAA .....((.(((((..((((((((..((((-(....))))).((((.((--------------((.(((((((.....)))))).).)))).))))))))))))..))))).))...... ( -37.90, z-score = -2.50, R) >droWil1.scaffold_181130 12693020 100 - 16660200 ------CUUGAUGUUCACACUUUACGAAA-GUUAGUUUUCAGAGGCCC------------GUAAGGUGCGAAAACUGUUUUCAACAUUGUCGCCUGAAGUGUGGCCCUCGUCAUCAGCG ------..(((((.(((((((((((....-))((((((((....((((------------....)).))))))))))..................)))))))))....)))))...... ( -28.00, z-score = -1.62, R) >consensus __UUUGUGUGAUGUUCACACUUUAGGAAA_GUUUGGUUUG_GGCCACG____CAGGGCC_GCGAGGUGCGAAAACA__________UUGUCGCCCGAAGUGUGGCCAUCAUCAUCAUGG ........(((((..((((((((..........((((.....))))..................((.((((.((............)).))))))))))))))..)))))......... (-23.23 = -22.76 + -0.47)
| Location | 15,189,217 – 15,189,317 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.46 |
| Shannon entropy | 0.55709 |
| G+C content | 0.51494 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15189217 100 - 27905053 CCAUGAUGAUGAUGGCCACACUUCGGGCGACAA----------UGUCUUCGCACCUGGC-GGCCCUG----CGUGGCC-CAAACCAAAC-UUUCCUAAAGUGUGAACAUCACACAAA-- ...((.((.(((((..(((((((..(((((...----------.....))))...(((.-((((...----...))))-....)))...-....)..)))))))..)))))))))..-- ( -32.20, z-score = -2.16, R) >droPer1.super_6 5022110 107 - 6141320 UGAUGAUGAUGAUGGCCACACUUGGGGCGACAA----------UGUUUUAGCACCUUACGGGCAGUGGGGCUGAGGCCGCAAACCAAACUUUUCGUAAAGUGUGAACAUCACUGGGA-- .........(((((..(((((((...((((.((----------.((((..((.((....))))..((.(((....))).))....)))))).)))).)))))))..)))))......-- ( -33.70, z-score = -1.21, R) >dp4.chr2 29651123 106 - 30794189 UGAUGAUGAUGAUGGCCACACUUGGGGCGACAA----------UGUUUUAGCACCUUACGGGCAGUGGGGCUGAGGCCGCAAACCAAAC-UUUCGUAAAGUGUGAACAUCACUGGGA-- .........(((((..(((((((((.(((....----------.((((((((.(((.((.....))))))))))))))))...))..((-....)).)))))))..)))))......-- ( -33.40, z-score = -1.14, R) >droAna3.scaffold_13340 8496596 96 + 23697760 --AUGAUGAUGAUGGCCACACUUCGGGCGACAA----------UGUUUUCGCACCUCCC-GGCCUC-----AGAGGGCCAAAACCAAAC-UUUCCUAAAGUGUGAACAUCACAAA---- --....((.(((((..(((((((..(((((...----------.....)))).......-(((((.-----...)))))..........-....)..)))))))..)))))))..---- ( -32.50, z-score = -3.33, R) >droEre2.scaffold_4770 11301127 99 + 17746568 CCAUGAUGAUGAUGGCCACACUUCGGGCGACAA----------UGUCUUCGCACCUCGC-GGCCCUG----CGC-ACC-CAAACCAAAC-UUUCCUAAAGUGUGAACAUCACACAAA-- ...((.((.(((((..(((((((..(((((...----------.....)))).....((-(......----)))-...-..........-....)..)))))))..)))))))))..-- ( -28.10, z-score = -2.43, R) >droYak2.chr3R 3911179 100 - 28832112 CCAUGAUGAUGAUGGCCACACUUCGGGCGACAA----------UGUCUUCGCACCUCGC-GGCCCUG----CGUGGUC-CAAACCAAAC-UUUCCUAAAGUGUGAACAUCACACAAG-- ...((.((.(((((..(((((((..(((((...----------.....))))(((.(((-(....))----)).))).-..........-....)..)))))))..)))))))))..-- ( -32.70, z-score = -2.74, R) >droSec1.super_25 248625 100 + 827797 CCAUGAUGAUGAUGGCCACACUUCGGGCGACAA----------UGUCUUCGCACCUCGC-GGCCCUG----CGUGGCC-CAAACCAAAC-UUUCCAAAAGUGUGAACAUCACACAAA-- ...((.(((((..((((((.(...(((((((..----------.)))..(((.....))-))))).)----.))))))-.....((.((-((.....)))).))..)))))))....-- ( -31.30, z-score = -2.20, R) >droSim1.chr3R 21227945 100 + 27517382 CCAUGAUGAUGAUGGCCACACUUCGGGCGACAA----------UGUCUUCGCACCUCGC-GGCCCUG----CGUGGCC-CAAACCAAAC-UUUCCUAAAGUGUGAACAUCACACAAA-- ...((.((.(((((..(((((((..(((((...----------(((....)))..))))-((((...----...))))-..........-....)..)))))))..)))))))))..-- ( -31.60, z-score = -2.50, R) >droVir3.scaffold_13047 2779955 116 + 19223366 --UCGGUGACGAUGGCCACACUUCGGGCGACAACGUUGCAAACUGUUUUCGCACCUCGCGGCCACAAAACAUUGGCCCUGAAAACCAAC-UUUCCUAAAGUGUGAACAUCAACCGGCAG --(((((...((((..(((((((..((((((...))))).....((((((((.....))(((((........)))))..))))))....-....)..)))))))..)))).)))))... ( -41.80, z-score = -3.81, R) >droMoj3.scaffold_6540 21387936 116 + 34148556 --UCGGUGACGAUGGCCACACUUUGGGCGACAACGUUGCAAACUGUUUUCGCACCUCCCGGCCAGAUAGCGACGGCCCUGAAAACCAAC-UUUCCUAAAGUGUGAACAUCAGCCUCACA --..(((...((((..((((((((((((((.((((........)))).))))...((..((((..........))))..))........-....))))))))))..)))).)))..... ( -37.70, z-score = -2.35, R) >droGri2.scaffold_15116 429877 104 + 1808639 UUUCGGUGAUGAUGGCCACACUUUGGGCGACAACACUGCAAACUGUUUUCGCACCUC--------------GCGGCCCUGAAAACCAAC-UUUCCUAAAGUGUGAACAUCAGCAAGUGG ...((.(..(((((..((((((((((((((.((((........)))).)))).....--------------........((((......-))))))))))))))..)))))...).)). ( -30.90, z-score = -1.52, R) >droWil1.scaffold_181130 12693020 100 + 16660200 CGCUGAUGACGAGGGCCACACUUCAGGCGACAAUGUUGAAAACAGUUUUCGCACCUUAC------------GGGCCUCUGAAAACUAAC-UUUCGUAAAGUGUGAACAUCAAG------ ..........((.(..(((((((...((((....(((......(((((((((.((....------------))))....))))))))))-..)))).)))))))..).))...------ ( -22.50, z-score = -0.06, R) >consensus CCAUGAUGAUGAUGGCCACACUUCGGGCGACAA__________UGUUUUCGCACCUCGC_GGCCCUG____CGGGGCC_CAAACCAAAC_UUUCCUAAAGUGUGAACAUCACACAAA__ ..........((((..(((((((.((((((..................)))).))..................((.........))...........)))))))..))))......... (-16.16 = -16.75 + 0.59)

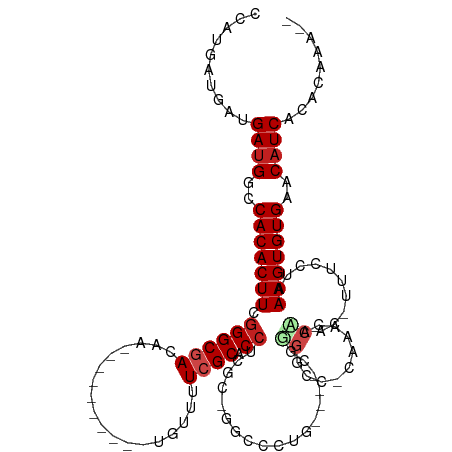
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:36 2011