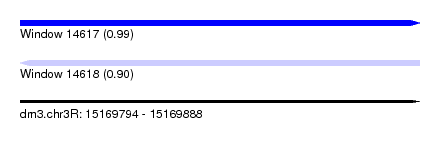
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,169,794 – 15,169,888 |
| Length | 94 |
| Max. P | 0.989978 |

| Location | 15,169,794 – 15,169,888 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 70.81 |
| Shannon entropy | 0.59058 |
| G+C content | 0.50207 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -17.91 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.989978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
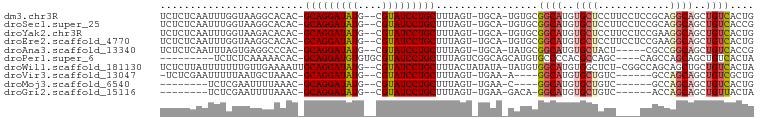
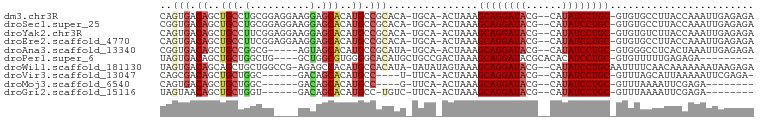
>dm3.chr3R 15169794 94 + 27905053 UCUCUCAAUUUGGUAAGGCACAC-GCAGGAUAUG--CGUAUCCUGCUUUAGU-UGCA-UGUGCGGCAUGUGCUCCUUCCUCCGCAGGCAGCUGUCACUG ...........((.((((..(((-((((((((..--..))))))))....((-(((.-...)))))..)))..))))))...((((....))))..... ( -30.30, z-score = -0.89, R) >droSec1.super_25 229051 94 - 827797 UCUCUCAAUUUGGUAAGGCACAC-GCAGGAUAUG--CGUAUCCUGCUUUAGU-UGCA-UGUGCGGCAUGUGCUCCUUCCUCCGCAGGCAGCUGUCACCG ...........(((..((((((.-((((((((..--..))))))))....((-(((.-...))))).)))))).........((((....)))).))). ( -31.10, z-score = -1.23, R) >droYak2.chr3R 3888650 94 + 28832112 UCUCUCAAUUUGGUAAGACACAC-GCAGGAUAUG--CGUAUCCUGCUUUAGU-UGCA-UGUGCGGCAUGUGCUCCUUCCUCCGAAGGCAGCUGUCACUG ................((((((.-((((((((..--..))))))))....((-(((.-...))))).)))((((((((....))))).))).))).... ( -31.50, z-score = -2.03, R) >droEre2.scaffold_4770 11281551 94 - 17746568 UCUCUCAAUUUGGUAAGGCACAC-GCAGGAUAUG--CGUAUCCUGCUUUAGU-UGCA-UGUGCGGCAUGUGCUCCUUCCUCCGAAGGCAGCUGUCACUG ........(((((.((((..(((-((((((((..--..))))))))....((-(((.-...)))))..)))..))))...)))))(((....))).... ( -31.70, z-score = -1.59, R) >droAna3.scaffold_13340 8478190 89 - 23697760 UCUCUCAAUUUAGUGAGGCCCAC-GCAGGAUAUG--CGUAUCCUGCUUUAGU-UGCA-UAUGCGGCAUGUGCUACU-----CGCCGGCAGCUGUCACCG ............(((((...(((-((((((((..--..))))))))....((-(((.-...)))))..)))...))-----))).(((....))).... ( -30.20, z-score = -1.32, R) >droPer1.super_6 5000772 85 + 6141320 ---------UCUCUCAAAAACAC-GCAGGAUGUGUGCGUAUCCUGCUUUAGUCGGCAGCAUGUGCCCCACGCCAGC----CAGCCAGCAGCUGUCACUA ---------..............-((((((((......))))))))..((((.((((((.((.((.....))))((----......)).)))))))))) ( -26.20, z-score = -0.83, R) >droWil1.scaffold_181130 12743390 95 - 16660200 UCUCUUAUUUUUUUGUUGAAAAUUGCAGGAUAUG--CGUAUCCUGCUUUACUAUAUA-UAUGUGGCAUGUGGCUCU-CGGCCAGCAGCUGCUGUCACUA ......((((((.....)))))).((((((((..--..))))))))...........-...((((((.(..(((((-.....)).)))..))))))).. ( -27.00, z-score = -1.90, R) >droVir3.scaffold_13047 2757831 83 - 19223366 -UCUCGAAUUUUUAAUGCUAAAC-GCAGGAUAUG--CGUAUCCUGCUUUAGU-UGAA-A----GGCAUGUGCUGUC------GCCAGCAGCUGUCGCUG -........(((((..((((((.-((((((((..--..))))))))))))))-))))-)----((((..(((((..------..)))))..)))).... ( -28.20, z-score = -2.38, R) >droMoj3.scaffold_6540 21365402 76 - 34148556 --------UCUCGAAUUUUAAAC-GCAGGAUAUG--CGUAUCCUGCUUUAGU-UGAA-C----GGCAUGUGCUGUC------GCCAGCAGCUGUCACUG --------..((((...(((((.-((((((((..--..))))))))))))))-))).-.----((((..(((((..------..)))))..)))).... ( -24.80, z-score = -2.06, R) >droGri2.scaffold_15116 406722 79 - 1808639 --------UCUCGAAUUUUAAAC-GCAGGAUAUG--CGUAUCCUGCUUUAGU-UGAA-GACA-GGCAUGUGCUGUC------ACCAGCAGCUGUUACUA --------............(((-((((((((..--..))))))))....((-((..-((((-(.(....))))))------..))))....))).... ( -23.50, z-score = -1.68, R) >consensus UCUCUCAAUUUGGUAAGGCACAC_GCAGGAUAUG__CGUAUCCUGCUUUAGU_UGCA_UAUGCGGCAUGUGCUCCU____CCGCCAGCAGCUGUCACUG ........................(((((((((....))))))))).................((((..((((............))))..)))).... (-17.91 = -18.28 + 0.37)
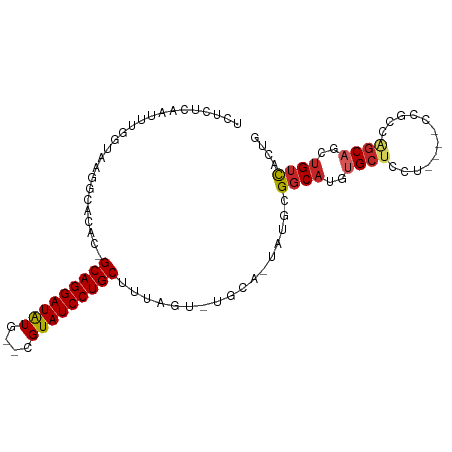
| Location | 15,169,794 – 15,169,888 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.81 |
| Shannon entropy | 0.59058 |
| G+C content | 0.50207 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.23 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.897440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15169794 94 - 27905053 CAGUGACAGCUGCCUGCGGAGGAAGGAGCACAUGCCGCACA-UGCA-ACUAAAGCAGGAUACG--CAUAUCCUGC-GUGUGCCUUACCAAAUUGAGAGA ((((....((.....))...((.(((.((((((...((...-.)).-......((((((((..--..))))))))-))))))))).))..))))..... ( -29.90, z-score = -1.24, R) >droSec1.super_25 229051 94 + 827797 CGGUGACAGCUGCCUGCGGAGGAAGGAGCACAUGCCGCACA-UGCA-ACUAAAGCAGGAUACG--CAUAUCCUGC-GUGUGCCUUACCAAAUUGAGAGA .(....).((.....))...((.(((.((((((...((...-.)).-......((((((((..--..))))))))-))))))))).))........... ( -30.60, z-score = -1.19, R) >droYak2.chr3R 3888650 94 - 28832112 CAGUGACAGCUGCCUUCGGAGGAAGGAGCACAUGCCGCACA-UGCA-ACUAAAGCAGGAUACG--CAUAUCCUGC-GUGUGUCUUACCAAAUUGAGAGA ((((....(((.(((((....))))))))((((((.((...-.)).-......((((((((..--..))))))))-))))))........))))..... ( -29.80, z-score = -1.57, R) >droEre2.scaffold_4770 11281551 94 + 17746568 CAGUGACAGCUGCCUUCGGAGGAAGGAGCACAUGCCGCACA-UGCA-ACUAAAGCAGGAUACG--CAUAUCCUGC-GUGUGCCUUACCAAAUUGAGAGA ((((....))))(((((....))))).((((((...((...-.)).-......((((((((..--..))))))))-))))))((((......))))... ( -30.70, z-score = -1.85, R) >droAna3.scaffold_13340 8478190 89 + 23697760 CGGUGACAGCUGCCGGCG-----AGUAGCACAUGCCGCAUA-UGCA-ACUAAAGCAGGAUACG--CAUAUCCUGC-GUGGGCCUCACUAAAUUGAGAGA .(....)....(((.((.-----(((.(((.(((...))).-))).-)))...((((((((..--..))))))))-)).)))((((......))))... ( -30.20, z-score = -1.29, R) >droPer1.super_6 5000772 85 - 6141320 UAGUGACAGCUGCUGGCUG----GCUGGCGUGGGGCACAUGCUGCCGACUAAAGCAGGAUACGCACACAUCCUGC-GUGUUUUUGAGAGA--------- .........(((((((.((----((.((((((.....)))))))))).))..)))))((((((((.......)))-))))).........--------- ( -29.40, z-score = -0.24, R) >droWil1.scaffold_181130 12743390 95 + 16660200 UAGUGACAGCAGCUGCUGGCCG-AGAGCCACAUGCCACAUA-UAUAUAGUAAAGCAGGAUACG--CAUAUCCUGCAAUUUUCAACAAAAAAAUAAGAGA ..(((...(((..((.((((..-...))))))))))))...-...........((((((((..--..))))))))........................ ( -21.50, z-score = -1.69, R) >droVir3.scaffold_13047 2757831 83 + 19223366 CAGCGACAGCUGCUGGC------GACAGCACAUGCC----U-UUCA-ACUAAAGCAGGAUACG--CAUAUCCUGC-GUUUAGCAUUAAAAAUUCGAGA- ...(((((..(((((..------..)))))..))..----.-....-.(((((((((((((..--..))))))))-.)))))..........)))...- ( -25.10, z-score = -2.36, R) >droMoj3.scaffold_6540 21365402 76 + 34148556 CAGUGACAGCUGCUGGC------GACAGCACAUGCC----G-UUCA-ACUAAAGCAGGAUACG--CAUAUCCUGC-GUUUAAAAUUCGAGA-------- .((((((.(((((((..------..)))))...)).----)-))..-)))...((((((((..--..))))))))-...............-------- ( -23.90, z-score = -2.15, R) >droGri2.scaffold_15116 406722 79 + 1808639 UAGUAACAGCUGCUGGU------GACAGCACAUGCC-UGUC-UUCA-ACUAAAGCAGGAUACG--CAUAUCCUGC-GUUUAAAAUUCGAGA-------- ((((.((((((((((..------..)))))...).)-))).-....-))))..((((((((..--..))))))))-...............-------- ( -24.40, z-score = -2.48, R) >consensus CAGUGACAGCUGCCGGCGG____AGGAGCACAUGCCGCACA_UGCA_ACUAAAGCAGGAUACG__CAUAUCCUGC_GUGUGCCUUACCAAAUUGAGAGA ((((....)))).........................................((((((((......))))))))........................ (-12.47 = -12.23 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:35 2011