| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,152,742 – 15,152,834 |
| Length | 92 |
| Max. P | 0.973083 |

| Location | 15,152,742 – 15,152,834 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.36742 |
| G+C content | 0.39943 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
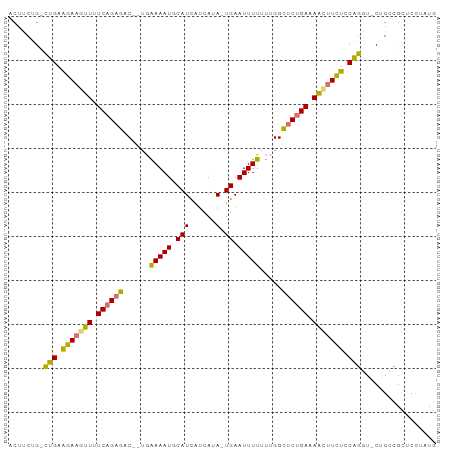
>dm3.chr3R 15152742 92 + 27905053 ACUUCUG-CUGAAGACGUUUUCAGAGAC--GGAAAAUGCAUCAUCAUA-UGAAUUUUUUUGGCUCUGAUAACUUCUCCAGGA-CUGUCGCUCUUAUG .......-(((.(((.(((.((((((.(--((((((..(((......)-))...))))))).)))))).))).))).)))..-.............. ( -22.80, z-score = -1.88, R) >droSim1.chr3R 21188117 92 - 27517382 ACUUCUG-UUGAAGAAGUUUUCAGAGAC--UGAAAAUGCAUCAUCAUA-UGAAUUUUUUUGGCUCUGAAAACUUCUCCAGGU-CUGCCGCUCCUAUG .......-.((.((((((((((((((.(--..((((....((((...)-)))...))))..))))))))))))))).))((.-...))......... ( -28.60, z-score = -3.92, R) >droSec1.super_25 212311 92 - 827797 ACUUCUG-CUGAAGAAAUUUUCAGAGAC--UGAAAAUGCAUCAUCAUA-UGAAUUUUGUUGGCUCUGAAAACUUCUCCAGGU-CUGCCGCUCCUAUG ......(-(((.((((.(((((((((.(--..(.......((((...)-)))......)..)))))))))).)))).))((.-...))))....... ( -21.12, z-score = -1.59, R) >droEre2.scaffold_4784 21223725 97 + 25762168 ACACCUGACAGAGAAAGAUUUCCGAGCCAAUGGAAAUGCAUUUGCAAAAUGAAUUUCAAGGGUCCAGACAUUUUUUUCUGCUUCUGCUGCUCUUCUU ........(((((((((((.((.(..((....(((((.((((.....)))).)))))...))..).)).)))))))))))................. ( -22.30, z-score = -0.54, R) >consensus ACUUCUG_CUGAAGAAGUUUUCAGAGAC__UGAAAAUGCAUCAUCAUA_UGAAUUUUUUUGGCUCUGAAAACUUCUCCAGGU_CUGCCGCUCCUAUG ........(((.((((((((((((((.....((((((.((.........)).))))))....)))))))))))))).)))................. (-18.41 = -18.23 + -0.19)


| Location | 15,152,742 – 15,152,834 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.36742 |
| G+C content | 0.39943 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.07 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
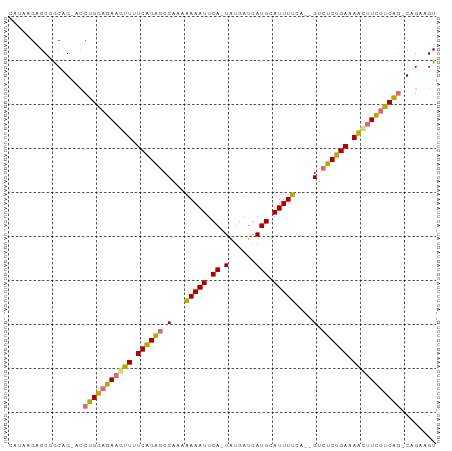
>dm3.chr3R 15152742 92 - 27905053 CAUAAGAGCGACAG-UCCUGGAGAAGUUAUCAGAGCCAAAAAAAUUCA-UAUGAUGAUGCAUUUUCC--GUCUCUGAAAACGUCUUCAG-CAGAAGU ..............-..(((((((.(((.((((((.(...(((((.((-(......))).)))))..--).)))))).))).)))))))-....... ( -21.30, z-score = -1.57, R) >droSim1.chr3R 21188117 92 + 27517382 CAUAGGAGCGGCAG-ACCUGGAGAAGUUUUCAGAGCCAAAAAAAUUCA-UAUGAUGAUGCAUUUUCA--GUCUCUGAAAACUUCUUCAA-CAGAAGU .........((...-.))(((((((((((((((((.(...(((((.((-(......))).)))))..--).))))))))))))))))).-....... ( -26.80, z-score = -2.73, R) >droSec1.super_25 212311 92 + 827797 CAUAGGAGCGGCAG-ACCUGGAGAAGUUUUCAGAGCCAACAAAAUUCA-UAUGAUGAUGCAUUUUCA--GUCUCUGAAAAUUUCUUCAG-CAGAAGU .......((((...-.))(((((((((((((((((.(...(((((.((-(......))).)))))..--).))))))))))))))))))-)...... ( -27.10, z-score = -2.61, R) >droEre2.scaffold_4784 21223725 97 - 25762168 AAGAAGAGCAGCAGAAGCAGAAAAAAAUGUCUGGACCCUUGAAAUUCAUUUUGCAAAUGCAUUUCCAUUGGCUCGGAAAUCUUUCUCUGUCAGGUGU .......(((.(....(((((.(((.((.((((..((..((((((.((((.....)))).)))).))..))..)))).)).))).)))))..).))) ( -19.60, z-score = 0.90, R) >consensus CAUAAGAGCGGCAG_ACCUGGAGAAGUUUUCAGAGCCAAAAAAAUUCA_UAUGAUGAUGCAUUUUCA__GUCUCUGAAAACUUCUUCAG_CAGAAGU .................((((((((((((((((((.(...(((((.((.........)).)))))....).))))))))))))))))))........ (-20.70 = -20.07 + -0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:32 2011