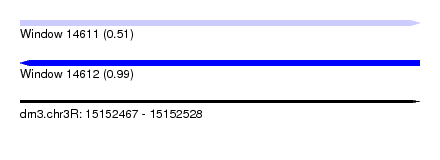
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,152,467 – 15,152,528 |
| Length | 61 |
| Max. P | 0.990975 |

| Location | 15,152,467 – 15,152,528 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 65.37 |
| Shannon entropy | 0.63717 |
| G+C content | 0.31526 |
| Mean single sequence MFE | -11.36 |
| Consensus MFE | -3.64 |
| Energy contribution | -4.64 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505088 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 15152467 61 + 27905053 AAGCAAAAUGCUUAGCAACAUUUAUUGUUGCCAAGAUGA---------GAACCAAAAAUAAAAUUAAAGA ((((.....)))).((((((.....))))))........---------...................... ( -10.00, z-score = -1.81, R) >droSim1.chr3R 21187877 69 - 27517382 AAGCAAAAUGCUUAGCAACAUUAAUUGUUGCCAAGAUAA-GUUGUUGAGAACCAAAAAUGAAAUUAAAGA .(((((....(((.((((((.....)))))).)))....-.)))))........................ ( -13.30, z-score = -2.36, R) >droYak2.chr3R 3871756 70 + 28832112 AAGCAAAAUGCUUAGCAACAUUUCUUGUUGAGAAGACGACUUUGAAGGGAACUCAAAAUCCAAUUUAGCA ..((.((((.(((..(((((.....)))))..)))..((.(((((.(....)))))).))..)))).)). ( -15.10, z-score = -1.55, R) >droEre2.scaffold_4770 11265311 70 - 17746568 AAGCGAAAUGUUUAGCAACAUUUCUCGUUGCCAAGACGAAGUUGAAAAUACCAGAAAAUUAAAUUUACGA .(((((((((((....))))))))(((((.....))))).)))........................... ( -11.30, z-score = -1.30, R) >droVir3.scaffold_13047 16473987 59 + 19223366 AAGGCAAUUGCCUAUCCAUAUCUAUUUCCCGCAAGAUGC-----AAACUAAAUAAAAAUGAAAA------ .((((....)))).................((.....))-----....................------ ( -7.10, z-score = -0.95, R) >consensus AAGCAAAAUGCUUAGCAACAUUUAUUGUUGCCAAGAUGA__UUGAAAAGAACCAAAAAUGAAAUUAAAGA ..........(((.((((((.....)))))).)))................................... ( -3.64 = -4.64 + 1.00)



| Location | 15,152,467 – 15,152,528 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 65.37 |
| Shannon entropy | 0.63717 |
| G+C content | 0.31526 |
| Mean single sequence MFE | -13.10 |
| Consensus MFE | -6.30 |
| Energy contribution | -7.22 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990975 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
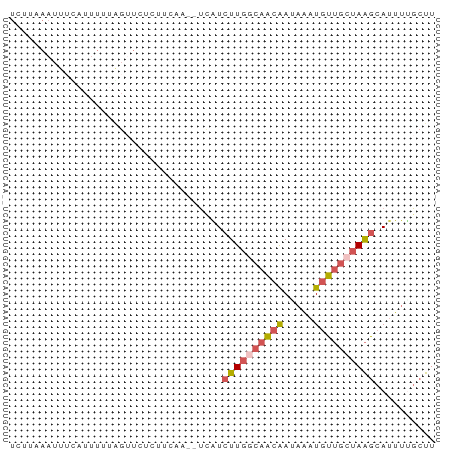
>dm3.chr3R 15152467 61 - 27905053 UCUUUAAUUUUAUUUUUGGUUC---------UCAUCUUGGCAACAAUAAAUGUUGCUAAGCAUUUUGCUU .................(((..---------....((((((((((.....))))))))))......))). ( -13.20, z-score = -2.65, R) >droSim1.chr3R 21187877 69 + 27517382 UCUUUAAUUUCAUUUUUGGUUCUCAACAAC-UUAUCUUGGCAACAAUUAAUGUUGCUAAGCAUUUUGCUU ..............................-....((((((((((.....)))))))))).......... ( -12.00, z-score = -2.11, R) >droYak2.chr3R 3871756 70 - 28832112 UGCUAAAUUGGAUUUUGAGUUCCCUUCAAAGUCGUCUUCUCAACAAGAAAUGUUGCUAAGCAUUUUGCUU .((.......(((((((((.....))))))))).............((((((((....)))))))))).. ( -14.60, z-score = -1.51, R) >droEre2.scaffold_4770 11265311 70 + 17746568 UCGUAAAUUUAAUUUUCUGGUAUUUUCAACUUCGUCUUGGCAACGAGAAAUGUUGCUAAACAUUUCGCUU ((((.............(((.....))).....((....)).))))((((((((....)))))))).... ( -11.50, z-score = -1.24, R) >droVir3.scaffold_13047 16473987 59 - 19223366 ------UUUUCAUUUUUAUUUAGUUU-----GCAUCUUGCGGGAAAUAGAUAUGGAUAGGCAAUUGCCUU ------..(((((.(((((((...((-----((.....)))).))))))).))))).((((....)))). ( -14.20, z-score = -2.22, R) >consensus UCUUAAAUUUCAUUUUUAGUUCUCUUCAA__UCAUCUUGGCAACAAUAAAUGUUGCUAAGCAUUUUGCUU ...................................((((((((((.....)))))))))).......... ( -6.30 = -7.22 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:30 2011