
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,139,842 – 15,139,934 |
| Length | 92 |
| Max. P | 0.966714 |

| Location | 15,139,842 – 15,139,934 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.72 |
| Shannon entropy | 0.35235 |
| G+C content | 0.44500 |
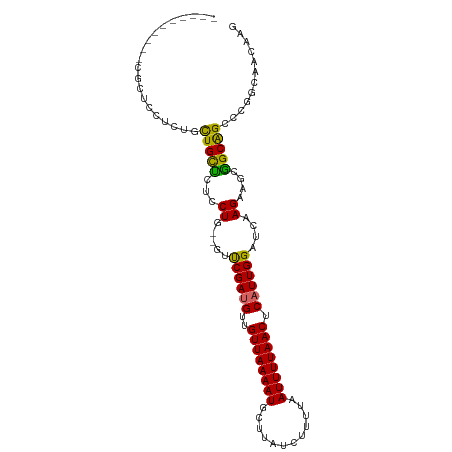
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -14.42 |
| Energy contribution | -13.78 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
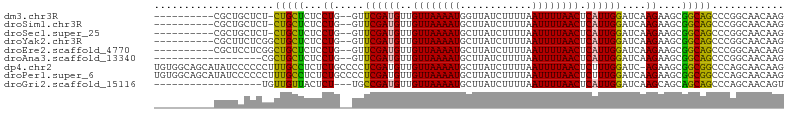
>dm3.chr3R 15139842 92 - 27905053 ----------CGCUGCUCU-CUGCUCUCCUG--GUUCGAUGUUGUUAAAAUGGUUAUCUUUUAAUUUUAACUCAUUGGAUCAAGAAGCGGCAGCCCGGCAACAAG ----------.((((..((-(((((.((.((--((((((((..((((((((............)))))))).)))))))))).))))))).))..))))...... ( -31.40, z-score = -4.10, R) >droSim1.chr3R 21175431 92 + 27517382 ----------CGCUGCUCU-CUGCUCUCCUG--GUUCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCAUUGGAUCAAGAAGCGGCAGCCCGGCAACAAG ----------.((((..((-(((((.((.((--((((((((..((((((((............)))))))).)))))))))).))))))).))..))))...... ( -31.40, z-score = -4.29, R) >droSec1.super_25 199436 92 + 827797 ----------CGCUGCUCU-CUGCUCUCCUG--GUUCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCAUUGGAUCAAGAAGCGGCAGCCCGGCAACAAG ----------.((((..((-(((((.((.((--((((((((..((((((((............)))))))).)))))))))).))))))).))..))))...... ( -31.40, z-score = -4.29, R) >droYak2.chr3R 3858617 93 - 28832112 ----------CGCUUCUCGGCUGCUCUCCUG--GUUCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCAUUGGAUCAAGAAGCGGCAGCCCGGCAACAAG ----------.(((....((((((((((.((--((((((((..((((((((............)))))))).)))))))))).).)).))))))).)))...... ( -32.60, z-score = -4.26, R) >droEre2.scaffold_4770 11252927 93 + 17746568 ----------CGCUCCUCGGCUGCUCUCCUG--GUUCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCAUUGGAUCAAGAAGCGGCAGCCCGGCAACAAG ----------.(((....((((((((((.((--((((((((..((((((((............)))))))).)))))))))).).)).))))))).)))...... ( -32.60, z-score = -4.19, R) >droAna3.scaffold_13340 8450034 85 + 23697760 ------------------CGCUGCUCUCCUG--GUUCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCAUUGGAUCAAGAAGCGGCAGCCCGGCAACAAG ------------------.((((((.((.((--((((((((..((((((((............)))))))).)))))))))).)))))))).....(....)... ( -29.50, z-score = -4.22, R) >dp4.chr2 29597416 104 - 30794189 UGUGGCAGCAUAUCCCCCCUUUGCCUCUCUGCCCCUCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCUUUGGAUC-AGAAGCGGCGGCCCAGCAACAAG (((.((.(((...........)))....((((((.(((((.(.((((((((............)))))))).....).)))-.)).).)))))....)).))).. ( -18.80, z-score = 0.92, R) >droPer1.super_6 4969517 105 - 6141320 UGUGGCAGCAUAUCCCCCCUUUGCCUCUCUGCCCCUCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCUUUGGAUCAAGAAGCGGCGGCCCAGCAACAAG ((.(((.(((...........)))..(.((((..((.(((.(.((((((((............)))))))).....).))).))..)))).))))))........ ( -18.50, z-score = 1.04, R) >droGri2.scaffold_15116 370573 84 + 1808639 ------------------UGUUGUUACUCU---UGCCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCAUUGGAUCAAGCAGCAGCAGCCCAGCAACAGU ------------------.((((((.((((---((((((((..((((((((............)))))))).))))))..)))).)).))))))........... ( -21.90, z-score = -2.20, R) >consensus __________CGCUCCUCUGCUGCUCUCCUG__GUUCGAUGUUGUUAAAAUGCUUAUCUUUUAAUUUUAACUCAUUGGAUCAAGAAGCGGCAGCCCGGCAACAAG ....................(((((...((.....((((((..((((((((............)))))))).))))))....))....)))))............ (-14.42 = -13.78 + -0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:28 2011