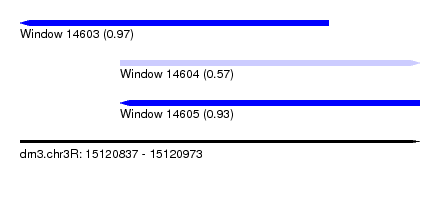
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,120,837 – 15,120,973 |
| Length | 136 |
| Max. P | 0.968865 |

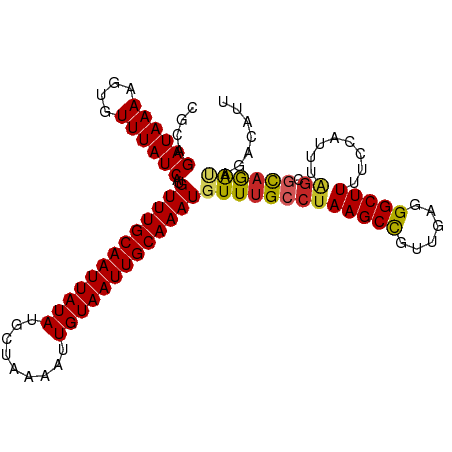
| Location | 15,120,837 – 15,120,942 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.36 |
| Shannon entropy | 0.21719 |
| G+C content | 0.35321 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -22.93 |
| Energy contribution | -22.73 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.968865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
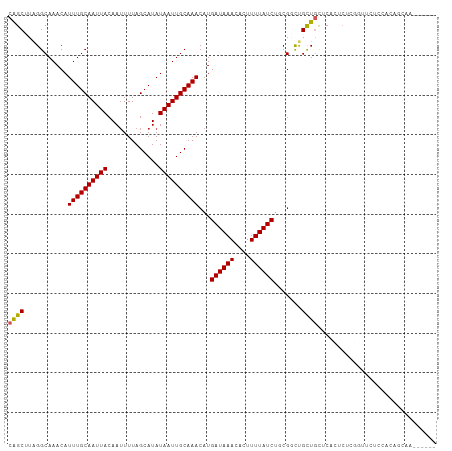
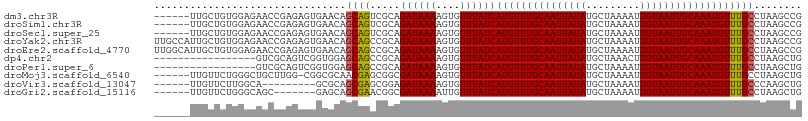
>dm3.chr3R 15120837 105 - 27905053 CGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCGUUGAGGGCUUUUCCAUUUAGCGCAGAUAGACAUU ..........(((((((((((((((((((((((((.........))))))))))))))).((((((((((......))))........))).))))))))))))) ( -31.40, z-score = -3.22, R) >droSim1.chr3R 21156319 105 + 27517382 CGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCGUUGAGGGCUUUUCCAUUUAGCGCAGAUAGACAUU ..........(((((((((((((((((((((((((.........))))))))))))))).((((((((((......))))........))).))))))))))))) ( -31.40, z-score = -3.22, R) >droSec1.super_25 180979 105 + 827797 CGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCGUUGAGGGCUUUUCCAUUUAGCGCAGAUAGACAUU ..........(((((((((((((((((((((((((.........))))))))))))))).((((((((((......))))........))).))))))))))))) ( -31.40, z-score = -3.22, R) >droYak2.chr3R 3832822 105 - 28832112 CGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCGUUGAGGGCUUUUCCAUUUAGCGCAGAUAGACAUU ..........(((((((((((((((((((((((((.........))))))))))))))).((((((((((......))))........))).))))))))))))) ( -31.40, z-score = -3.22, R) >droEre2.scaffold_4770 11234439 105 + 17746568 CGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCGUUGAGGGCUUUUCCAUUUAGCGCAGAUAGACAUU ..........(((((((((((((((((((((((((.........))))))))))))))).((((((((((......))))........))).))))))))))))) ( -31.40, z-score = -3.22, R) >droAna3.scaffold_13340 8431990 105 + 23697760 AACAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUGAGGGCUUUUCCAUUUGGCGCAGAUAGACGUU .............((((((((((((((((((((((.........)))))))))))))))..(((.(((((......)))))........)))...)))))))... ( -28.10, z-score = -1.95, R) >dp4.chr2 29576364 102 - 30794189 CGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAACUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUGAGGGCUUUUCCAUUUAGCGUAGA-AGCCA-- .(((((((((....))))))(((((((((((((((.........))))))))))))))).))).............(((((((.(.......).)))-)))).-- ( -26.60, z-score = -1.53, R) >droPer1.super_6 4948393 102 - 6141320 CGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUGAGGGCUUUUCCAUUUAGCGUAGA-AGCCA-- .(((((((((....))))))(((((((((((((((.........))))))))))))))).))).............(((((((.(.......).)))-)))).-- ( -26.60, z-score = -1.68, R) >droVir3.scaffold_13047 2704644 103 + 19223366 CGGAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCCAAGCUGUUAAGGGCUUUACU-CGCAGC-UAAACCUUCAAC .((.((((((....))))))(((((((((((((((.........)))))))))))))))....)).((((((...(((.....))-))))))-)........... ( -26.50, z-score = -2.13, R) >droGri2.scaffold_15116 346024 105 + 1808639 CGGCGAUAAAAUUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUAAAGGGCUUUACUCUCAGAUACUGGCACACACA ....((((((....))))))..(((((((((((((.........)))))))))))))((.((((....(((...((((.....))))))).....)))).))... ( -27.40, z-score = -2.20, R) >consensus CGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCGUUGAGGGCUUUUCCAUUUAGCGCAGAUAGACAUU ....((((((....))))))..(((((((((((((.........)))))))))))))(((((((((((((......)))))........)).))))))....... (-22.93 = -22.73 + -0.20)

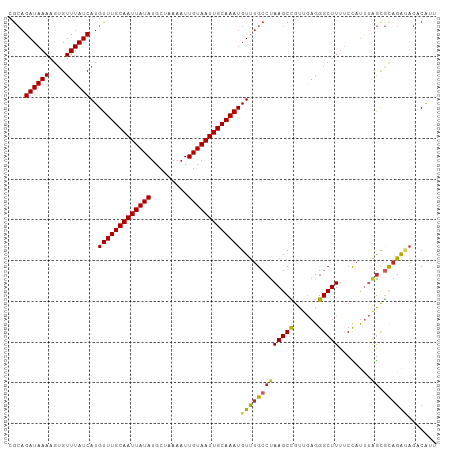
| Location | 15,120,871 – 15,120,973 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Shannon entropy | 0.37395 |
| G+C content | 0.39274 |
| Mean single sequence MFE | -20.44 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.32 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566013 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
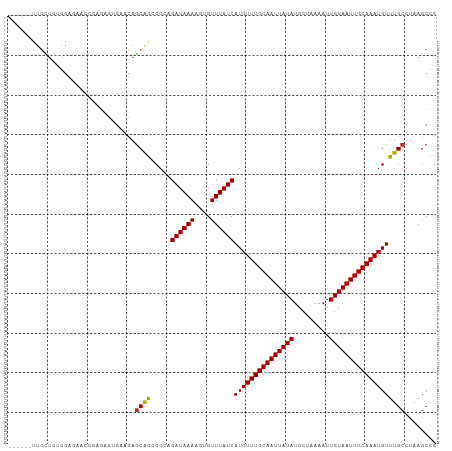
>dm3.chr3R 15120871 102 + 27905053 CGGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGACUGCUGUUCACUCUCGGUUCUCCACAGCAA------ ..(((...(((....((((((((((.............))))))))))...((((((....)))))))))((..((((........))))..))...)))..------ ( -18.92, z-score = -1.13, R) >droSim1.chr3R 21156353 102 - 27517382 CGGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGACUGCUGUUCACUCUCGGUUCUCCACAGCAA------ ..(((...(((....((((((((((.............))))))))))...((((((....)))))))))((..((((........))))..))...)))..------ ( -18.92, z-score = -1.13, R) >droSec1.super_25 181013 102 - 827797 CGGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGACUGCUGUUCACUCUCGGUUCUCCACAGCAA------ ..(((...(((....((((((((((.............))))))))))...((((((....)))))))))((..((((........))))..))...)))..------ ( -18.92, z-score = -1.13, R) >droYak2.chr3R 3832856 108 + 28832112 CGGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGGCUGCUGUUCACUCUCGGUUCUCCACAGCAAUGGCAA ..(((...((.....((((((((((.............))))))))))...((((((....))))))((.((..((((........))))...)).))))...))).. ( -22.02, z-score = -0.74, R) >droEre2.scaffold_4770 11234473 108 - 17746568 CGGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGGCUGCUGUUCACUCUCGGUUCUCCACAGCAAUGCCAA .(((....(((....((((((((((.............))))))))))...((((((....)))))))))...((((((........((....))))))))..))).. ( -24.22, z-score = -1.67, R) >dp4.chr2 29576395 91 + 30794189 CAGCUUAGGCAAACAUUUGCAAUUACAAGUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGGCUGCUCCACCGACUGCGAC----------------- (((((...(((....((((((((((...(.....)...))))))))))...((((((....))))))))))))))................----------------- ( -17.90, z-score = -0.79, R) >droPer1.super_6 4948424 91 + 6141320 CAGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGGCUGCUCCACCGACUGCGAC----------------- (((((...(((....((((((((((.............))))))))))...((((((....))))))))))))))................----------------- ( -17.32, z-score = -1.16, R) >droMoj3.scaffold_6540 21311424 101 - 34148556 CAGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCGCCGCUCGUUGCGCCG-CCAAGCAGCCCAGAACAA------ ..((((.(((.....((((((((((.............))))))))))...((((((....))))))(.(((.....))).))-))))))............------ ( -24.22, z-score = -2.25, R) >droVir3.scaffold_13047 2704676 93 - 19223366 CAGCUUGGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUCCGCUCGCUGCGC---------UGCCAAGAACAA------ ((((.(((((.....((((((((((.............))))))))))...((((((....))))))...)))))....))---------))..........------ ( -19.42, z-score = -1.19, R) >droGri2.scaffold_15116 346058 95 - 1808639 CAGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACAAUUUUAUCGCCGUUCGCUGCUC-------GCUGCCCAGAACAA------ .......(((.....((((((((((.............))))))))))...((((((....)))))))))((((...((..-------...))...))))..------ ( -22.52, z-score = -2.43, R) >consensus CAGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGGCUGCUGCUCACUCUCGGUUCUCCACAGCAA______ ((((...........((((((((((.............))))))))))...((((((....)))))).......)))).............................. (-13.46 = -13.32 + -0.14)

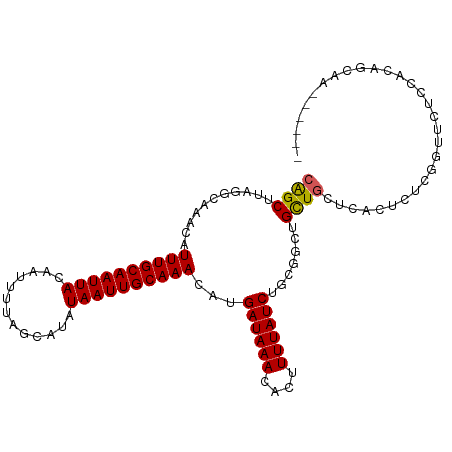
| Location | 15,120,871 – 15,120,973 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.87 |
| Shannon entropy | 0.37395 |
| G+C content | 0.39274 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -18.97 |
| Energy contribution | -18.65 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926831 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
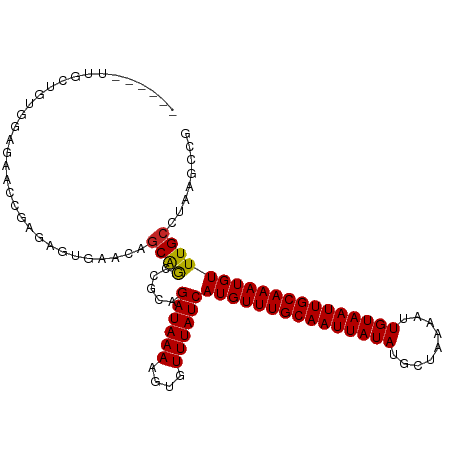
>dm3.chr3R 15120871 102 - 27905053 ------UUGCUGUGGAGAACCGAGAGUGAACAGCAGUCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCG ------(((((((.....((.....))..)))))))..(((((((((....))))))(((((((((((((((.........))))))))))))))).)))........ ( -26.10, z-score = -1.81, R) >droSim1.chr3R 21156353 102 + 27517382 ------UUGCUGUGGAGAACCGAGAGUGAACAGCAGUCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCG ------(((((((.....((.....))..)))))))..(((((((((....))))))(((((((((((((((.........))))))))))))))).)))........ ( -26.10, z-score = -1.81, R) >droSec1.super_25 181013 102 + 827797 ------UUGCUGUGGAGAACCGAGAGUGAACAGCAGUCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCG ------(((((((.....((.....))..)))))))..(((((((((....))))))(((((((((((((((.........))))))))))))))).)))........ ( -26.10, z-score = -1.81, R) >droYak2.chr3R 3832856 108 - 28832112 UUGCCAUUGCUGUGGAGAACCGAGAGUGAACAGCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCG .(((..(((((((.....((.....))..)))))))..)))((((((....))))))(((((((((((((((.........)))))))))))))))............ ( -27.50, z-score = -1.58, R) >droEre2.scaffold_4770 11234473 108 + 17746568 UUGGCAUUGCUGUGGAGAACCGAGAGUGAACAGCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCG ..(((.(((((((.....((.....))..)))))))..(((((((((....))))))(((((((((((((((.........))))))))))))))).)))....))). ( -32.00, z-score = -2.55, R) >dp4.chr2 29576395 91 - 30794189 -----------------GUCGCAGUCGGUGGAGCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAACUUGUAAUUGCAAAUGUUUGCCUAAGCUG -----------------....((((...(((.((((.....((((((....))))))(((((((((((((((.........)))))))))))))))))))))).)))) ( -24.50, z-score = -1.17, R) >droPer1.super_6 4948424 91 - 6141320 -----------------GUCGCAGUCGGUGGAGCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUG -----------------....((((...(((.((((.....((((((....))))))(((((((((((((((.........)))))))))))))))))))))).)))) ( -24.50, z-score = -1.43, R) >droMoj3.scaffold_6540 21311424 101 + 34148556 ------UUGUUCUGGGCUGCUUGG-CGGCGCAACGAGCGGCGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUG ------..((((((.(((((...)-)))).))..))))(((((((((....))))))(((((((((((((((.........)))))))))))))))..)))....... ( -34.00, z-score = -2.63, R) >droVir3.scaffold_13047 2704676 93 + 19223366 ------UUGUUCUUGGCA---------GCGCAGCGAGCGGAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCCAAGCUG ------..........((---------((...((((((...((((((....))))))..(((((((((((((.........)))))))))))))))))))....)))) ( -28.20, z-score = -2.19, R) >droGri2.scaffold_15116 346058 95 + 1808639 ------UUGUUCUGGGCAGC-------GAGCAGCGAACGGCGAUAAAAUUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUG ------..........((((-------.....((((((...((((((....))))))..(((((((((((((.........)))))))))))))))))))....)))) ( -28.10, z-score = -2.11, R) >consensus ______UUGCUGUGGAGAACCGAGAGUGAACAGCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCG ................................((((.....((((((....))))))(((((((((((((((.........)))))))))))))))))))........ (-18.97 = -18.65 + -0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:24 2011