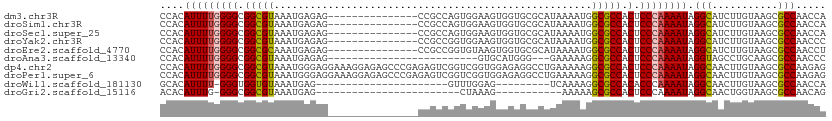
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,113,646 – 15,113,742 |
| Length | 96 |
| Max. P | 0.976147 |

| Location | 15,113,646 – 15,113,742 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.91 |
| Shannon entropy | 0.48949 |
| G+C content | 0.54153 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.71 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
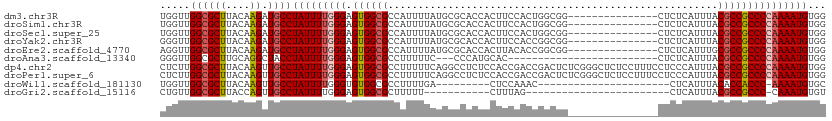
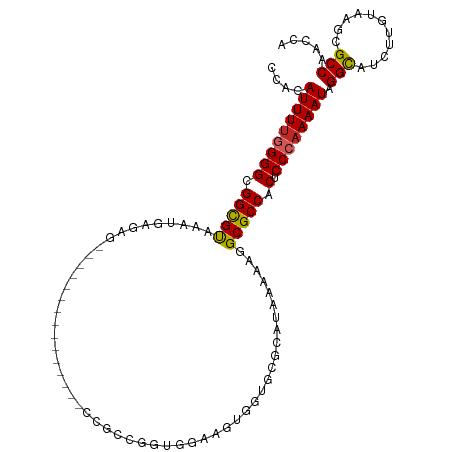
>dm3.chr3R 15113646 96 + 27905053 UGGUUGGCGCUUACAAGAUGCCUAUUUUGGGAGUGGCGCCAUUUUAUGCGCACCACUUCCACUGGCGG---------------CUCUCAUUUACGCCGCCCCAAAAUGUGG (((..((((((....)).)))).....(((((((((.(((((...))).)).)))))))))..(((((---------------(..........)))))))))........ ( -36.30, z-score = -2.30, R) >droSim1.chr3R 21149292 96 - 27517382 UGGUUGGCGCUUACAAGAUGCCUAUUUUGGGAGUGGCGCCAUUUUAUGCGCACCACUUCCACUGGCGG---------------CUCUCAUUUACGCCGCCCCAAAAUGUGG (((..((((((....)).)))).....(((((((((.(((((...))).)).)))))))))..(((((---------------(..........)))))))))........ ( -36.30, z-score = -2.30, R) >droSec1.super_25 174021 96 - 827797 UGGUUGGCGCUUACAAGAUGCCUAUUUUGGGAGUGGCGCCAUUUUAUGCGCACCACUUCCACUGGCGG---------------CUCUCAUUUACGCCGCCCCAAAAUGUGG (((..((((((....)).)))).....(((((((((.(((((...))).)).)))))))))..(((((---------------(..........)))))))))........ ( -36.30, z-score = -2.30, R) >droYak2.chr3R 3825112 96 + 28832112 GGGUUGGCGCUUACAAGAUGCCUAUUUUGGGAGUGGCGCCAUUUUAUGCGCACCACUUCCACCGGCGG---------------CUCUCAUUUACGCCGCCCCAAAAUGUGG (((.(((((......(((.(((.....(((((((((.(((((...))).)).))))))))).....))---------------))))......))))).)))......... ( -38.80, z-score = -2.67, R) >droEre2.scaffold_4770 11227121 96 - 17746568 AGGUUGGCGCUUACAAGAUGCCUAUUUUGGGAGUGGCGCCAUUUUAUGCGCACCACUUACACCGGCGG---------------CUCUCAUUUGCGCCGCCCCAAAAUGUGG .....((((((....)).))))((((((((((((((.(((((...))).)).)))))).....(((((---------------(.(......).))))))))))))))... ( -37.60, z-score = -2.20, R) >droAna3.scaffold_13340 8424766 83 - 23697760 GGGUUGGCGCUUGCAGGCUACCUAUUUUGGGAGUGGCGCCUUUUUC---CCCAUGCAC-------------------------CUCUCAUUUACGCCGCCCCAAAAUGUGG .(((..((....))..))).(((((((((((.((((((........---.........-------------------------..........))))))))))))))).)) ( -25.73, z-score = -0.06, R) >dp4.chr2 29569842 111 + 30794189 CUCUUGGCGCUUACAAGUUGCCUAUUUUGGGAGUGGCGCCUUUUUCAGGCCUCUCCACCGACCGACUCUCGGGCUCUCCUUUCCUCCCAUUUACGCCGCCCCAAAAUGUGG .....((((((....)).))))(((((((((.(((((((((.....)))).......((((.......))))......................))))))))))))))... ( -34.60, z-score = -2.09, R) >droPer1.super_6 4941874 111 + 6141320 CUCUUGGCGCUUACAAGUUGCCUAUUUUGGGAGUGGCGCCUUUUUCAGGCCUCUCCACCGACCGACUCUCGGGCUCUCCUUUCCUCCCAUUUACGCCGCCCCAAAAUGUGG .....((((((....)).))))(((((((((.(((((((((.....)))).......((((.......))))......................))))))))))))))... ( -34.60, z-score = -2.09, R) >droWil1.scaffold_181130 3858949 79 + 16660200 UGGUUGGCGCUUACAAGUUGCCUAUUUUGGGUGUGGCGCCUUUUGA---------CUCCAAAC----------------------CUCAUUUACACCACCC-AAAAUGUGC .....((((((....)).))))(((((((((((((......((((.---------...)))).----------------------........)).)))))-))))))... ( -19.06, z-score = -0.01, R) >droGri2.scaffold_15116 335973 75 - 1808639 CUGUUGGCGCUUACCAGUUGCCUAUUUUGGGAGUGGCGCUUUUU-----------CUUUAG------------------------CUCAUUUACGCCGCCC-CAAAUGUGU .....((((((....)).))))...((((((.((((((......-----------......------------------------........))))))))-))))..... ( -21.75, z-score = -1.22, R) >consensus UGGUUGGCGCUUACAAGAUGCCUAUUUUGGGAGUGGCGCCAUUUUAUGCGCACCACUUCCACCGGCGG_______________CUCUCAUUUACGCCGCCCCAAAAUGUGG .....((((((....)).))))(((((((((.((((((.......................................................)))))))))))))))... (-21.39 = -21.71 + 0.32)
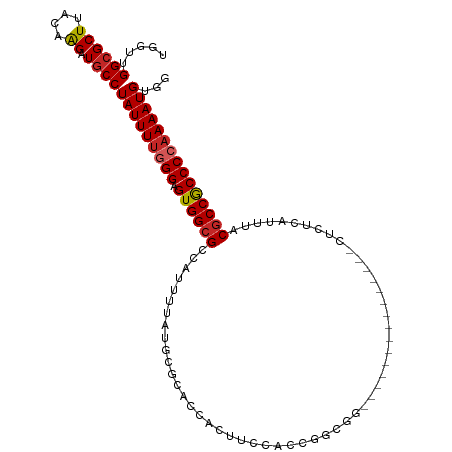
| Location | 15,113,646 – 15,113,742 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.91 |
| Shannon entropy | 0.48949 |
| G+C content | 0.54153 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -18.78 |
| Energy contribution | -18.81 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15113646 96 - 27905053 CCACAUUUUGGGGCGGCGUAAAUGAGAG---------------CCGCCAGUGGAAGUGGUGCGCAUAAAAUGGCGCCACUCCCAAAAUAGGCAUCUUGUAAGCGCCAACCA ..........((..(((((..(..((((---------------((.....(((.((((((((.(((...))))))))))).))).....))).)))..)..)))))..)). ( -42.50, z-score = -3.71, R) >droSim1.chr3R 21149292 96 + 27517382 CCACAUUUUGGGGCGGCGUAAAUGAGAG---------------CCGCCAGUGGAAGUGGUGCGCAUAAAAUGGCGCCACUCCCAAAAUAGGCAUCUUGUAAGCGCCAACCA ..........((..(((((..(..((((---------------((.....(((.((((((((.(((...))))))))))).))).....))).)))..)..)))))..)). ( -42.50, z-score = -3.71, R) >droSec1.super_25 174021 96 + 827797 CCACAUUUUGGGGCGGCGUAAAUGAGAG---------------CCGCCAGUGGAAGUGGUGCGCAUAAAAUGGCGCCACUCCCAAAAUAGGCAUCUUGUAAGCGCCAACCA ..........((..(((((..(..((((---------------((.....(((.((((((((.(((...))))))))))).))).....))).)))..)..)))))..)). ( -42.50, z-score = -3.71, R) >droYak2.chr3R 3825112 96 - 28832112 CCACAUUUUGGGGCGGCGUAAAUGAGAG---------------CCGCCGGUGGAAGUGGUGCGCAUAAAAUGGCGCCACUCCCAAAAUAGGCAUCUUGUAAGCGCCAACCC .........(((..(((((..(..((((---------------((.....(((.((((((((.(((...))))))))))).))).....))).)))..)..)))))..))) ( -44.10, z-score = -3.88, R) >droEre2.scaffold_4770 11227121 96 + 17746568 CCACAUUUUGGGGCGGCGCAAAUGAGAG---------------CCGCCGGUGUAAGUGGUGCGCAUAAAAUGGCGCCACUCCCAAAAUAGGCAUCUUGUAAGCGCCAACCU .........(((..(((((..(..((((---------------((.....((..((((((((.(((...)))))))))))..)).....))).)))..)..)))))..))) ( -41.70, z-score = -3.06, R) >droAna3.scaffold_13340 8424766 83 + 23697760 CCACAUUUUGGGGCGGCGUAAAUGAGAG-------------------------GUGCAUGGG---GAAAAAGGCGCCACUCCCAAAAUAGGUAGCCUGCAAGCGCCAACCC .........(((..(((((....(((.(-------------------------((((.....---.......))))).)))......((((...))))...)))))..))) ( -25.20, z-score = 0.36, R) >dp4.chr2 29569842 111 - 30794189 CCACAUUUUGGGGCGGCGUAAAUGGGAGGAAAGGAGAGCCCGAGAGUCGGUCGGUGGAGAGGCCUGAAAAAGGCGCCACUCCCAAAAUAGGCAACUUGUAAGCGCCAAGAG .....((((((.((........((((((....((...(((......((((((........)))).))....))).)).))))))....((....)).....)).)))))). ( -35.30, z-score = -0.44, R) >droPer1.super_6 4941874 111 - 6141320 CCACAUUUUGGGGCGGCGUAAAUGGGAGGAAAGGAGAGCCCGAGAGUCGGUCGGUGGAGAGGCCUGAAAAAGGCGCCACUCCCAAAAUAGGCAACUUGUAAGCGCCAAGAG .....((((((.((........((((((....((...(((......((((((........)))).))....))).)).))))))....((....)).....)).)))))). ( -35.30, z-score = -0.44, R) >droWil1.scaffold_181130 3858949 79 - 16660200 GCACAUUUU-GGGUGGUGUAAAUGAG----------------------GUUUGGAG---------UCAAAAGGCGCCACACCCAAAAUAGGCAACUUGUAAGCGCCAACCA ........(-((.((((((...((.(----------------------((.(((.(---------((....))).))).)))))....((....)).....)))))).))) ( -24.30, z-score = -1.05, R) >droGri2.scaffold_15116 335973 75 + 1808639 ACACAUUUG-GGGCGGCGUAAAUGAG------------------------CUAAAG-----------AAAAAGCGCCACUCCCAAAAUAGGCAACUGGUAAGCGCCAACAG .....((((-(((.(((((.......------------------------......-----------.....))))).).))))))...(((..((....)).)))..... ( -20.17, z-score = -1.07, R) >consensus CCACAUUUUGGGGCGGCGUAAAUGAGAG_______________CCGCCGGUGGAAGUGGUGCGCAUAAAAAGGCGCCACUCCCAAAAUAGGCAUCUUGUAAGCGCCAACCA ....(((((((((.(((((.....................................................))))).).)))))))).(((...........)))..... (-18.78 = -18.81 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:22 2011