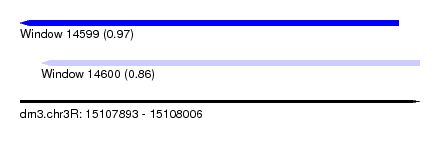
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,107,893 – 15,108,006 |
| Length | 113 |
| Max. P | 0.968035 |

| Location | 15,107,893 – 15,108,000 |
|---|---|
| Length | 107 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Shannon entropy | 0.43935 |
| G+C content | 0.39237 |
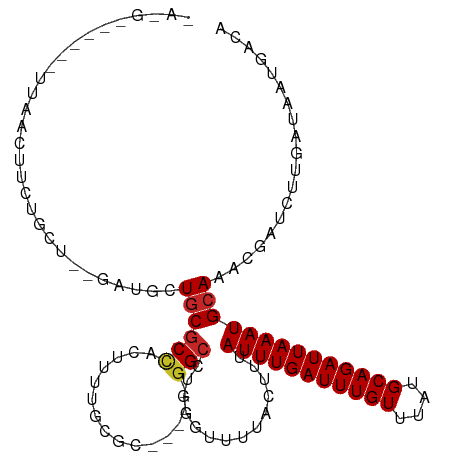
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -11.34 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.968035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
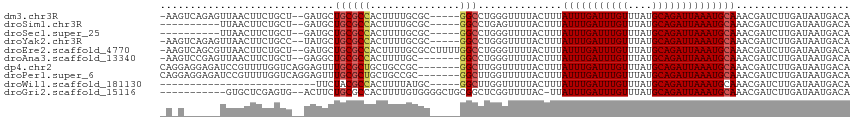
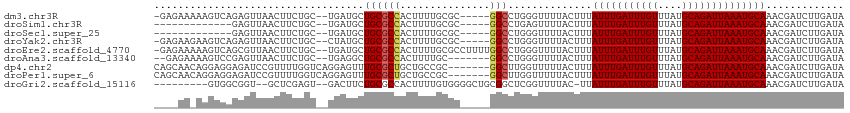
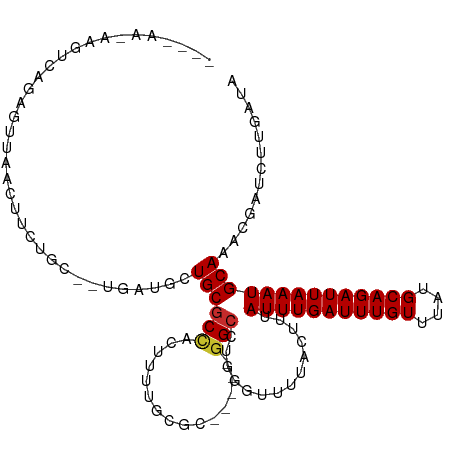
>dm3.chr3R 15107893 107 - 27905053 -AAGUCAGAGUUAACUUCUGCU--GAUGCUGCGCCACUUUUGCGC-----GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA -..((((..(((((..((.(((--.(.(((((((.......))))-----))).).))).......((((((((((((....)))))))))))).....))..)))))..)))). ( -33.50, z-score = -3.62, R) >droSim1.chr3R 21143354 98 + 27517382 ----------UUAACUUCUGCU--GAUGCUGCGCCACUUUUGCGC-----GGCCUGAGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA ----------((((..((.(((--.(.(((((((.......))))-----))).).))).......((((((((((((....)))))))))))).....))..))))........ ( -25.10, z-score = -3.01, R) >droSec1.super_25 168447 98 + 827797 ----------UUAACUUCUGCU--GAUGCUGCGCCACUUUUGCGC-----GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA ----------((((..((.(((--.(.(((((((.......))))-----))).).))).......((((((((((((....)))))))))))).....))..))))........ ( -25.10, z-score = -2.62, R) >droYak2.chr3R 3819537 107 - 28832112 -AAGUCAGAGUUAACUUCUGCC--UAUGCUGCGCCACUUUUGCGC-----GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA -..((((..(((((..((.(((--((.(((((((.......))))-----))).))))).......((((((((((((....)))))))))))).....))..)))))..)))). ( -37.40, z-score = -5.12, R) >droEre2.scaffold_4770 11221536 112 + 17746568 -AAGUCAGCGUUAACUUCUGCU--GAUGCUGCGCCACUUUUGCGCCUUUUGGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA -..((((..(((((..((.((.--......))......((((((((....)))..............(((((((((((....)))))))))))))))).))..)))))..)))). ( -28.00, z-score = -1.64, R) >droAna3.scaffold_13340 8418945 105 + 23697760 -AAGUCCGAGUUAACUUCUGCU--GAGGCUGCGCCACUUUUGC-------GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA -..(((...(((((..((.(((--.((((((((.......)))-------))))).))).......((((((((((((....)))))))))))).....))..)))))...))). ( -31.70, z-score = -3.29, R) >dp4.chr2 29564112 108 - 30794189 CAGGAGGAGAUCCGUUUUGGUCAGGAGUUUGCGCUGCUGCCGC-------GGCUUGGUUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA ..(((.....)))(((...((((((((((((((((((....))-------)))..............(((((((((((....)))))))))))))))))..)))))))...))). ( -37.30, z-score = -3.82, R) >droPer1.super_6 4936155 108 - 6141320 CAGGAGGAGAUCCGUUUUGGUCAGGAGUUUGCGCUGCUGCCGC-------GGCUUGGUUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA ..(((.....)))(((...((((((((((((((((((....))-------)))..............(((((((((((....)))))))))))))))))..)))))))...))). ( -37.30, z-score = -3.82, R) >droWil1.scaffold_181130 3852532 84 - 16660200 --------------------------UUCUACGCCACUUUUAUGC-----GGCUUGGUUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA --------------------------..(((.(((.(......).-----))).))).........((((((((((((....))))))))))))..................... ( -15.60, z-score = -1.78, R) >droGri2.scaffold_15116 330238 101 + 1808639 -----------GUGCUCGAGUG--ACUUCUGCGCCACUUUUGUGGGGCUGCGGCUCGGUUUUAC-UUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA -----------.(((((((((.--.(....((.((((....)))).)).)..))))))......-..(((((((((((....))))))))))))))................... ( -28.10, z-score = -1.90, R) >consensus _A_G______UUAACUUCUGCU__GAUGCUGCGCCACUUUUGCGC_____GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUAAUGACA .............................((((((...............)))..............(((((((((((....))))))))))))))................... (-11.34 = -11.50 + 0.16)
| Location | 15,107,899 – 15,108,006 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Shannon entropy | 0.44924 |
| G+C content | 0.40976 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -11.81 |
| Energy contribution | -11.88 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15107899 107 - 27905053 -GAGAAAAAGUCAGAGUUAACUUCUGC--UGAUGCUGCGCCACUUUUGCGC-----GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA -((((((((.(((((((........))--)...(((((((.......))))-----))))))).))))....((((((((((((....)))))))))))).......)))).... ( -29.50, z-score = -2.11, R) >droSim1.chr3R 21143360 95 + 27517382 -------------GAGUUAACUUCUGC--UGAUGCUGCGCCACUUUUGCGC-----GGCCUGAGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA -------------..(((((..((.((--(.(.(((((((.......))))-----))).).))).......((((((((((((....)))))))))))).....))..))))). ( -26.90, z-score = -3.29, R) >droSec1.super_25 168453 95 + 827797 -------------GAGUUAACUUCUGC--UGAUGCUGCGCCACUUUUGCGC-----GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA -------------..(((((..((.((--(.(.(((((((.......))))-----))).).))).......((((((((((((....)))))))))))).....))..))))). ( -26.90, z-score = -2.93, R) >droYak2.chr3R 3819543 107 - 28832112 -GAGAAGAAGUCAGAGUUAACUUCUGC--CUAUGCUGCGCCACUUUUGCGC-----GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA -((((((((((........))))))((--(((.(((((((.......))))-----))).))))).......((((((((((((....)))))))))))).......)))).... ( -34.20, z-score = -3.56, R) >droEre2.scaffold_4770 11221542 112 + 17746568 -GAGAAAAAGUCAGCGUUAACUUCUGC--UGAUGCUGCGCCACUUUUGCGCCUUUUGGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA -((((....(.((((((((.(....).--)))))))))....(.((((((((....)))..............(((((((((((....)))))))))))))))).).)))).... ( -29.00, z-score = -1.76, R) >droAna3.scaffold_13340 8418951 104 + 23697760 --GAGAAAAGUCCGAGUUAACUUCUGC--UGAGGCUGCGCCACUUUUGC-------GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA --(((((((..((.(((........))--).((((((((.......)))-------)))))))..)))....((((((((((((....)))))))))))).......)))).... ( -28.40, z-score = -1.96, R) >dp4.chr2 29564118 108 - 30794189 CAGCAACAGGAGGAGAUCCGUUUUGGUCAGGAGUUUGCGCUGCUGCCGC-------GGCUUGGUUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA ......(((((((....)).)))))((((((((((((((((((....))-------)))..............(((((((((((....)))))))))))))))))..))))))). ( -35.30, z-score = -2.82, R) >droPer1.super_6 4936161 108 - 6141320 CAGCAACAGGAGGAGAUCCGUUUUGGUCAGGAGUUUGCGCUGCUGCCGC-------GGCUUGGUUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA ......(((((((....)).)))))((((((((((((((((((....))-------)))..............(((((((((((....)))))))))))))))))..))))))). ( -35.30, z-score = -2.82, R) >droGri2.scaffold_15116 330244 101 + 1808639 ---------GUGGCGGU--GCUCGAGU--GACUUCUGCGCCACUUUUGUGGGGCUGCGGCUCGGUUUUAC-UUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA ---------..((((.(--((((((((--..(....((.((((....)))).)).)..))))))......-..(((((((((((....))))))))))))))..)).))...... ( -28.90, z-score = -1.29, R) >consensus ____AA_AAGUCAGAGUUAACUUCUGC__UGAUGCUGCGCCACUUUUGCGC_____GGCCUGGGUUUUACUUUAUUUGAUUUGUUUAUGCAGAUUAAAUGCAAACGAUCUUGAUA ...................................((((((...............)))..............(((((((((((....))))))))))))))............. (-11.81 = -11.88 + 0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:20 2011