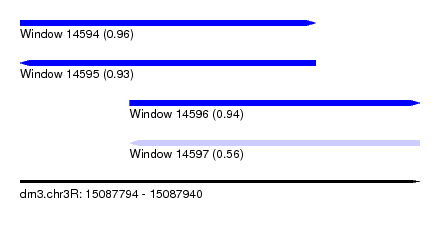
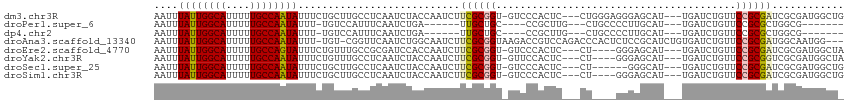
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 15,087,794 – 15,087,940 |
| Length | 146 |
| Max. P | 0.958013 |

| Location | 15,087,794 – 15,087,902 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.21 |
| Shannon entropy | 0.47400 |
| G+C content | 0.47616 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.81 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
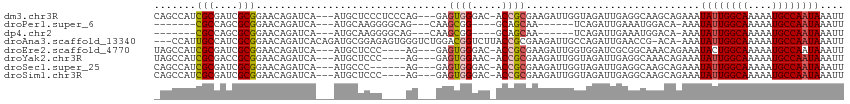
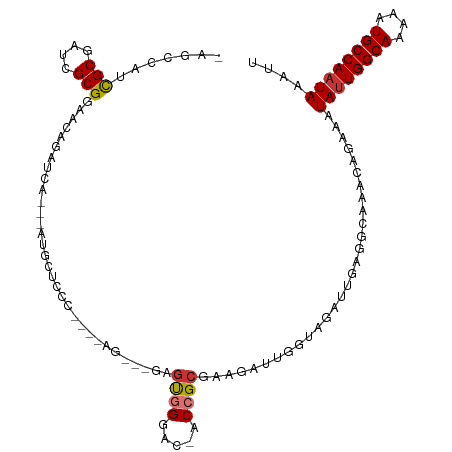
>dm3.chr3R 15087794 108 + 27905053 CAGCCAUCGCGAUCGCGGAACAGAUCA---AUGCUCCCUCCCAG---GAGUGGGAC-ACCGCGAAGAUUGGUAGAUUGAGGCAAGCAGAAAUAUUGGCAAAAAUGCCAAUAAAUU ..(((.((((((((........)))).---.((..(((((....---))).))..)-)..)))).((((....))))..))).........((((((((....)))))))).... ( -34.00, z-score = -2.28, R) >droPer1.super_6 4907665 91 + 6141320 -------CGCCAGCGCGGAACAGAUCA---AUGCAAGGGGCAG---CAAGCGG----GCAGCAA------UCAGAUUGAAAUGGACA-AAAUAUUGGCAAAAAUGCCAAUAAAUU -------(((....)))(..((..(((---((.(..(..((.(---(......----)).))..------)..))))))..))..).-...((((((((....)))))))).... ( -20.10, z-score = -0.30, R) >dp4.chr2 29536979 91 + 30794189 -------CGCCAGCGCGGAACAGAUCA---AUGCAAGGGGCAG---CAAGCGG----GCAGCAA------UCAGAUUGAAAUGGACA-AAAUAUUGGCAAAAAUGCCAAUAAAUU -------(((....)))(..((..(((---((.(..(..((.(---(......----)).))..------)..))))))..))..).-...((((((((....)))))))).... ( -20.10, z-score = -0.30, R) >droAna3.scaffold_13340 8398126 110 - 23697760 ---CCAUUGCCAUCGCGGAACAGAUCACAGAUGCGGAGAGUGGGUCUGGACGGUCUUACCGCGAAGAUUGCCAGAUUGAACCG-ACA-AAAUAUUGGCAAAAAUGCCAAUAAAUU ---.......((((...((.....))...))))(((....(.(((((((.(((((((......)))))))))))))).).)))-...-...((((((((....)))))))).... ( -32.90, z-score = -2.23, R) >droEre2.scaffold_4770 11202283 104 - 17746568 UAGCCAUCGCGAUCGCGGAACAGAUCA---AUGCUCCC----AG---GAGUGGGAC-ACCGCGAAGAUUGGUGGAUCGCGGCAAACAGAAAUACUGGCAAAAAUGCCAAUAAAUU ..(((.((((....))))....((((.---....((((----(.---...)))))(-((((.......)))))))))..)))............(((((....)))))....... ( -32.10, z-score = -1.93, R) >droYak2.chr3R 3797360 104 + 28832112 UAGCCAUCGCGACCGCGGAACAGAUCA---AUGCUCCC----AG---GAGUGGAAC-ACCGCGAAGAUUGGUAGAUUGAGGCAAACAGAAAUAUUGGCAAAAAUGCCAAUAAAUU ..(((.((((....))))......(((---((.((.((----((---..((((...-.)))).....)))).)))))))))).........((((((((....)))))))).... ( -32.90, z-score = -3.81, R) >droSec1.super_25 148594 102 - 827797 CAGCCAUCGCGAUCGCGGAACAGAUCA---AUGCCC------AG---GAGUGGGAC-ACCGCGAAGAUUGGUAGAUUGAGGCAAGCAGAAAUAUUGGCAAAAAUGCCAAUAAAUU ..(((.((((....))))......(((---((..((------((---..((((...-.)))).....))))...)))))))).........((((((((....)))))))).... ( -31.60, z-score = -2.83, R) >droSim1.chr3R 21123329 104 - 27517382 CAGCCAUCGCGAUCGCGGAACAGAUCA---AUGCUCCC----AG---GAGUGGGAC-ACCGCGAAGAUUGGUAGAUUGAGGCAAGCAGAAAUAUUGGCAAAAAUGCCAAUAAAUU ..(((.((((....))))......(((---((.((.((----((---..((((...-.)))).....)))).)))))))))).........((((((((....)))))))).... ( -34.20, z-score = -3.16, R) >consensus _AGCCAUCGCGAUCGCGGAACAGAUCA___AUGCUCCC____AG___GAGUGGGAC_ACCGCGAAGAUUGGUAGAUUGAGGCAAACAGAAAUAUUGGCAAAAAUGCCAAUAAAUU .......(((....)))................................((((.....)))).............................((((((((....)))))))).... (-15.65 = -15.81 + 0.16)
| Location | 15,087,794 – 15,087,902 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.21 |
| Shannon entropy | 0.47400 |
| G+C content | 0.47616 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.86 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15087794 108 - 27905053 AAUUUAUUGGCAUUUUUGCCAAUAUUUCUGCUUGCCUCAAUCUACCAAUCUUCGCGGU-GUCCCACUC---CUGGGAGGGAGCAU---UGAUCUGUUCCGCGAUCGCGAUGGCUG ....((((((((....)))))))).........(((...............(((((((-.(((((...---.))))).((((((.---.....))))))...))))))).))).. ( -35.79, z-score = -2.63, R) >droPer1.super_6 4907665 91 - 6141320 AAUUUAUUGGCAUUUUUGCCAAUAUUU-UGUCCAUUUCAAUCUGA------UUGCUGC----CCGCUUG---CUGCCCCUUGCAU---UGAUCUGUUCCGCGCUGGCG------- ((..((((((((....))))))))..)-)(..((..((((((...------..((.((----......)---).)).....).))---)))..))..)(((....)))------- ( -19.60, z-score = -0.71, R) >dp4.chr2 29536979 91 - 30794189 AAUUUAUUGGCAUUUUUGCCAAUAUUU-UGUCCAUUUCAAUCUGA------UUGCUGC----CCGCUUG---CUGCCCCUUGCAU---UGAUCUGUUCCGCGCUGGCG------- ((..((((((((....))))))))..)-)(..((..((((((...------..((.((----......)---).)).....).))---)))..))..)(((....)))------- ( -19.60, z-score = -0.71, R) >droAna3.scaffold_13340 8398126 110 + 23697760 AAUUUAUUGGCAUUUUUGCCAAUAUUU-UGU-CGGUUCAAUCUGGCAAUCUUCGCGGUAAGACCGUCCAGACCCACUCUCCGCAUCUGUGAUCUGUUCCGCGAUGGCAAUGG--- ....((((((((....))))))))..(-(((-(((......)))))))((.((((((..(((...((((((..(.......)..)))).)))))...)))))).))......--- ( -33.10, z-score = -2.46, R) >droEre2.scaffold_4770 11202283 104 + 17746568 AAUUUAUUGGCAUUUUUGCCAGUAUUUCUGUUUGCCGCGAUCCACCAAUCUUCGCGGU-GUCCCACUC---CU----GGGAGCAU---UGAUCUGUUCCGCGAUCGCGAUGGCUA ....((((((((....)))))))).........(((((((((((((.........)))-)........---..----.((((((.---.....))))))..))))))...))).. ( -34.00, z-score = -2.36, R) >droYak2.chr3R 3797360 104 - 28832112 AAUUUAUUGGCAUUUUUGCCAAUAUUUCUGUUUGCCUCAAUCUACCAAUCUUCGCGGU-GUUCCACUC---CU----GGGAGCAU---UGAUCUGUUCCGCGGUCGCGAUGGCUA ....((((((((....)))))))).........(((...............(((((((-(((((.(..---..----))))))))---)((((........)))))))).))).. ( -30.19, z-score = -2.35, R) >droSec1.super_25 148594 102 + 827797 AAUUUAUUGGCAUUUUUGCCAAUAUUUCUGCUUGCCUCAAUCUACCAAUCUUCGCGGU-GUCCCACUC---CU------GGGCAU---UGAUCUGUUCCGCGAUCGCGAUGGCUG ....((((((((....)))))))).........(((...............(((((((-(.((((...---.)------))))))---)((((........)))))))).))).. ( -28.99, z-score = -2.50, R) >droSim1.chr3R 21123329 104 + 27517382 AAUUUAUUGGCAUUUUUGCCAAUAUUUCUGCUUGCCUCAAUCUACCAAUCUUCGCGGU-GUCCCACUC---CU----GGGAGCAU---UGAUCUGUUCCGCGAUCGCGAUGGCUG ....((((((((....)))))))).........(((...............(((((((-((((((...---.)----)))).)))---)((((........)))))))).))).. ( -30.19, z-score = -2.33, R) >consensus AAUUUAUUGGCAUUUUUGCCAAUAUUUCUGUUUGCCUCAAUCUACCAAUCUUCGCGGU_GUCCCACUC___CU____GGGAGCAU___UGAUCUGUUCCGCGAUCGCGAUGGCU_ ....((((((((....))))))))...........................((((((........................................))))))............ (-14.74 = -14.86 + 0.13)
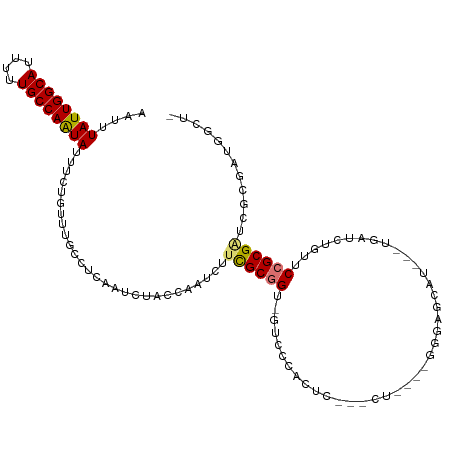
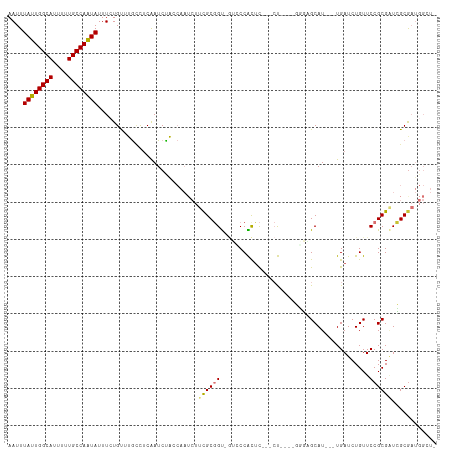
| Location | 15,087,834 – 15,087,940 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.31361 |
| G+C content | 0.35309 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -12.15 |
| Energy contribution | -12.39 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15087834 106 + 27905053 -----------GGAGUGGGACACCGCGAAGAUUGGUAGAUUGAGGCAAGCAGAAAUAUUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUAACUUUAUUAAAU-- -----------...((((....))))((((.((((((...(((......((((((((((((((....))))))))..))))))........)))......)))))))))).......-- ( -23.94, z-score = -3.00, R) >droPer1.super_6 4907691 105 + 6141320 -----------GGGGCAGCAAGCGGGCAGCAAU--CAGAUUGAAAUGGACA-AAAUAUUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUGUCUUUAUUAAAAAC -----------(((((((((.((.....))...--..........((..((-(((((((((((....))))))))...))))).))..............))))))))).......... ( -25.00, z-score = -2.83, R) >dp4.chr2 29537005 105 + 30794189 -----------GGGGCAGCAAGCGGGCAGCAAU--CAGAUUGAAAUGGACA-AAAUAUUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUGUCUUUAUUAAAAAC -----------(((((((((.((.....))...--..........((..((-(((((((((((....))))))))...))))).))..............))))))))).......... ( -25.00, z-score = -2.83, R) >droAna3.scaffold_13340 8398159 115 - 23697760 AGAGUGGGUCUGGACGGUCUUACCGCGAAGAUUGCCAGAUUGAACCGA--CAAAAUAUUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUAACUUUAUUAAAC-- (((((.(((((((.(((((((......)))))))))))))).......--(((((((((((((....))))))))...)))))...((.((((.....)))))).))))).......-- ( -30.10, z-score = -3.82, R) >droEre2.scaffold_4770 11202319 106 - 17746568 -----------GGAGUGGGACACCGCGAAGAUUGGUGGAUCGCGGCAAACAGAAAUACUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUAACUUUAUUAAAU-- -----------...((((..(((((.......)))))..))))((((..(((((((..(((((....)))))....)))))))....(........)...)))).............-- ( -22.90, z-score = -2.12, R) >droYak2.chr3R 3797396 106 + 28832112 -----------GGAGUGGAACACCGCGAAGAUUGGUAGAUUGAGGCAAACAGAAAUAUUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUAACUUUAUUAAAU-- -----------...((((....))))((((.((((((..(((.......((((((((((((((....))))))))..))))))..((....))...))).)))))))))).......-- ( -22.20, z-score = -2.99, R) >droSec1.super_25 148628 106 - 827797 -----------GGAGUGGGACACCGCGAAGAUUGGUAGAUUGAGGCAAGCAGAAAUAUUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUAACUUUAUUAAAU-- -----------...((((....))))((((.((((((...(((......((((((((((((((....))))))))..))))))........)))......)))))))))).......-- ( -23.94, z-score = -3.00, R) >droSim1.chr3R 21123365 106 - 27517382 -----------GGAGUGGGACACCGCGAAGAUUGGUAGAUUGAGGCAAGCAGAAAUAUUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUAACUUUAUUAAAU-- -----------...((((....))))((((.((((((...(((......((((((((((((((....))))))))..))))))........)))......)))))))))).......-- ( -23.94, z-score = -3.00, R) >consensus ___________GGAGUGGGACACCGCGAAGAUUGGUAGAUUGAGGCAAACAGAAAUAUUGGCAAAAAUGCCAAUAAAUUUUUGACAAGACAUUAUACAAAUGCUAACUUUAUUAAAU__ ...........(((((((...............................((((((((((((((....))))))))..))))))......((((.....))))))).))))......... (-12.15 = -12.39 + 0.23)

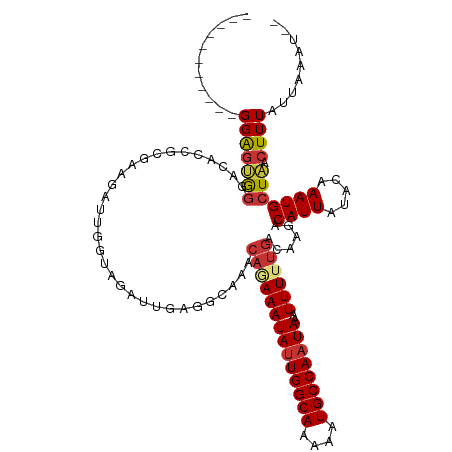
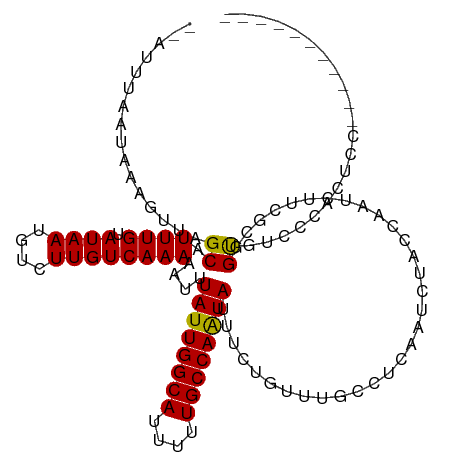
| Location | 15,087,834 – 15,087,940 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.31361 |
| G+C content | 0.35309 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -12.02 |
| Energy contribution | -11.73 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 15087834 106 - 27905053 --AUUUAAUAAAGUUAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAAUAUUUCUGCUUGCCUCAAUCUACCAAUCUUCGCGGUGUCCCACUCC----------- --..........((.((((((((.((((....))))))))....((((((((....))))))))....)))).)).......((((.........)))).........----------- ( -17.90, z-score = -1.95, R) >droPer1.super_6 4907691 105 - 6141320 GUUUUUAAUAAAGACAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAAUAUUU-UGUCCAUUUCAAUCUG--AUUGCUGCCCGCUUGCUGCCCC----------- ............(((((((((.....)))))..))))...((..((((((((....))))))))..)-)...............--...((.((......)).))...----------- ( -19.70, z-score = -1.20, R) >dp4.chr2 29537005 105 - 30794189 GUUUUUAAUAAAGACAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAAUAUUU-UGUCCAUUUCAAUCUG--AUUGCUGCCCGCUUGCUGCCCC----------- ............(((((((((.....)))))..))))...((..((((((((....))))))))..)-)...............--...((.((......)).))...----------- ( -19.70, z-score = -1.20, R) >droAna3.scaffold_13340 8398159 115 + 23697760 --GUUUAAUAAAGUUAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAAUAUUUUG--UCGGUUCAAUCUGGCAAUCUUCGCGGUAAGACCGUCCAGACCCACUCU --((((.(((((.......))))).....(((((..(.......((((((((....))))))))..(((--((((......))))))).......)..))))).....))))....... ( -25.60, z-score = -2.28, R) >droEre2.scaffold_4770 11202319 106 + 17746568 --AUUUAAUAAAGUUAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAGUAUUUCUGUUUGCCGCGAUCCACCAAUCUUCGCGGUGUCCCACUCC----------- --..............(((((.....))))).............((((((((....)))))))).....(..((((((((...........))))))))..)......----------- ( -23.30, z-score = -2.95, R) >droYak2.chr3R 3797396 106 - 28832112 --AUUUAAUAAAGUUAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAAUAUUUCUGUUUGCCUCAAUCUACCAAUCUUCGCGGUGUUCCACUCC----------- --..........((.((((((((.((((....))))))))....((((((((....))))))))....)))).)).......((((.........)))).........----------- ( -15.50, z-score = -1.18, R) >droSec1.super_25 148628 106 + 827797 --AUUUAAUAAAGUUAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAAUAUUUCUGCUUGCCUCAAUCUACCAAUCUUCGCGGUGUCCCACUCC----------- --..........((.((((((((.((((....))))))))....((((((((....))))))))....)))).)).......((((.........)))).........----------- ( -17.90, z-score = -1.95, R) >droSim1.chr3R 21123365 106 + 27517382 --AUUUAAUAAAGUUAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAAUAUUUCUGCUUGCCUCAAUCUACCAAUCUUCGCGGUGUCCCACUCC----------- --..........((.((((((((.((((....))))))))....((((((((....))))))))....)))).)).......((((.........)))).........----------- ( -17.90, z-score = -1.95, R) >consensus __AUUUAAUAAAGUUAGCAUUUGUAUAAUGUCUUGUCAAAAAUUUAUUGGCAUUUUUGCCAAUAUUUCUGUUUGCCUCAAUCUACCAAUCUUCGCGGUGUCCCACUCC___________ ................((.((((.((((....))))))))....((((((((....))))))))................................))..................... (-12.02 = -11.73 + -0.30)

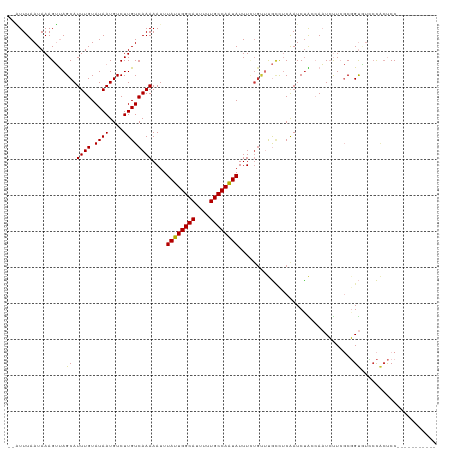
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:27:18 2011